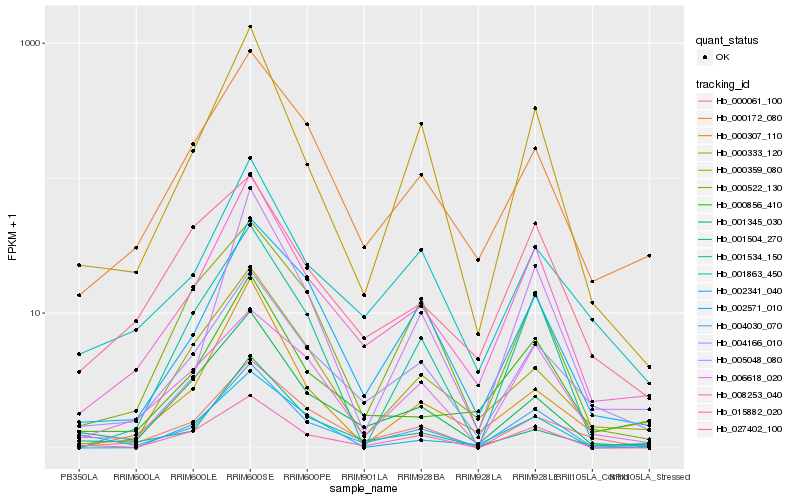
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008253_040 |
0.0 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 2 |
Hb_000856_410 |
0.1066322401 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_004166_010 |
0.1182802935 |
- |
- |
PREDICTED: uncharacterized protein LOC105638508 [Jatropha curcas] |
| 4 |
Hb_000172_080 |
0.1218217007 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 5 |
Hb_004030_070 |
0.1316328468 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 6 |
Hb_001863_450 |
0.1378336946 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 7 |
Hb_001345_030 |
0.1429937292 |
- |
- |
hypothetical protein POPTR_0007s10390g, partial [Populus trichocarpa] |
| 8 |
Hb_000333_120 |
0.1432069363 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |
| 9 |
Hb_002571_010 |
0.145581647 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 10 |
Hb_000061_100 |
0.152475766 |
- |
- |
hypothetical protein CISIN_1g0053742mg, partial [Citrus sinensis] |
| 11 |
Hb_001504_270 |
0.1571627258 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 12 |
Hb_000359_080 |
0.161220566 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 13 |
Hb_015882_020 |
0.1628903026 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 14 |
Hb_027402_100 |
0.1681605998 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |
| 15 |
Hb_005048_080 |
0.1727812473 |
- |
- |
PREDICTED: PAP-specific phosphatase HAL2-like [Jatropha curcas] |
| 16 |
Hb_001534_150 |
0.1731053579 |
- |
- |
PREDICTED: uncharacterized permease C29B12.14c [Jatropha curcas] |
| 17 |
Hb_002341_040 |
0.1746267786 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 18 |
Hb_006618_020 |
0.1750660758 |
- |
- |
Vacuolar-processing enzyme precursor [Ricinus communis] |
| 19 |
Hb_000522_130 |
0.1766544851 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 20 |
Hb_000307_110 |
0.1767754826 |
- |
- |
hypothetical protein JCGZ_08160 [Jatropha curcas] |