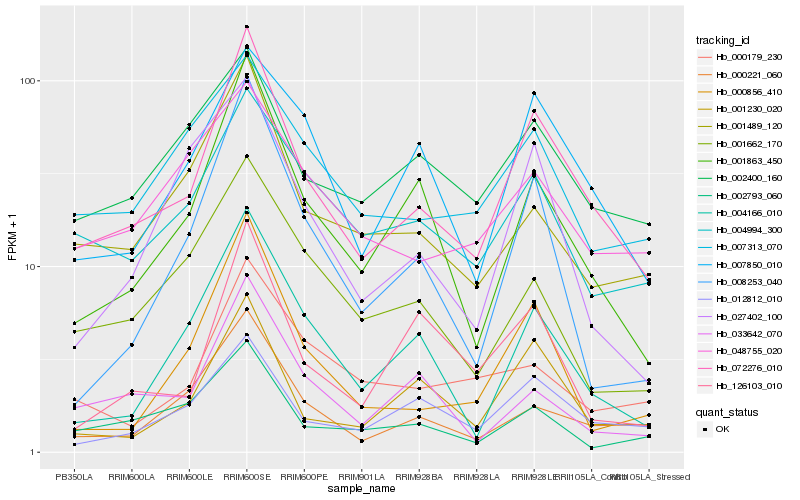
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_072276_010 |
0.0 |
- |
- |
PREDICTED: elongation of fatty acids protein 3-like [Jatropha curcas] |
| 2 |
Hb_001863_450 |
0.1481140622 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 3 |
Hb_001230_020 |
0.1519399453 |
- |
- |
hypothetical protein POPTR_0001s31610g [Populus trichocarpa] |
| 4 |
Hb_012812_010 |
0.1578544492 |
- |
- |
putative leucine-rich repeat receptor-like serine/threonine-protein kinase [Morus notabilis] |
| 5 |
Hb_000221_060 |
0.1648709176 |
- |
- |
- |
| 6 |
Hb_007850_010 |
0.1655890296 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.4 [Jatropha curcas] |
| 7 |
Hb_004166_010 |
0.1689485343 |
- |
- |
PREDICTED: uncharacterized protein LOC105638508 [Jatropha curcas] |
| 8 |
Hb_000856_410 |
0.1699716207 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_004994_300 |
0.1704768556 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 10 |
Hb_001489_120 |
0.1739588493 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002400_160 |
0.1811652933 |
- |
- |
Ubiquitin-protein ligase BRE1A, putative [Ricinus communis] |
| 12 |
Hb_008253_040 |
0.1816902863 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 13 |
Hb_126103_010 |
0.1853449728 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 14 |
Hb_007313_070 |
0.1868498513 |
- |
- |
PREDICTED: gibberellin receptor GID1C [Jatropha curcas] |
| 15 |
Hb_033642_070 |
0.1883521323 |
- |
- |
PREDICTED: oligopeptide transporter 7-like [Jatropha curcas] |
| 16 |
Hb_002793_060 |
0.1907119291 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g02980 [Populus euphratica] |
| 17 |
Hb_001662_170 |
0.1930278097 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 18 |
Hb_000179_230 |
0.1936896595 |
- |
- |
PREDICTED: myosin-17 [Jatropha curcas] |
| 19 |
Hb_027402_100 |
0.1939185044 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |
| 20 |
Hb_048755_020 |
0.1947204507 |
- |
- |
hypothetical protein JCGZ_09860 [Jatropha curcas] |