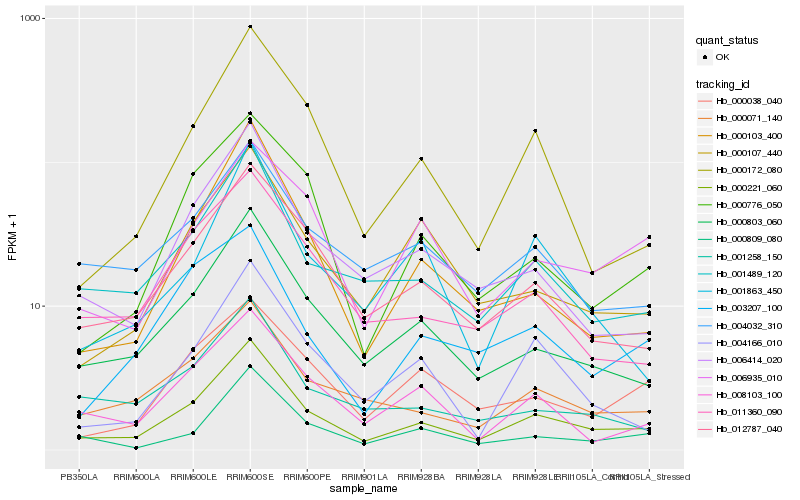
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000221_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_000803_060 |
0.1132831908 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 3 |
Hb_001489_120 |
0.1245730403 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_012787_040 |
0.1305796838 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000071_140 |
0.1350185632 |
- |
- |
PREDICTED: uncharacterized protein LOC105628258 [Jatropha curcas] |
| 6 |
Hb_006414_020 |
0.1398452264 |
transcription factor |
TF Family: MYB |
- |
| 7 |
Hb_000103_400 |
0.1451554031 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000172_080 |
0.1457992928 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 9 |
Hb_000809_080 |
0.1461569795 |
- |
- |
50S ribosomal protein L25, putative [Ricinus communis] |
| 10 |
Hb_008103_100 |
0.1482759177 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPS2 [Jatropha curcas] |
| 11 |
Hb_004166_010 |
0.1514279991 |
- |
- |
PREDICTED: uncharacterized protein LOC105638508 [Jatropha curcas] |
| 12 |
Hb_011360_090 |
0.1542510009 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 13 |
Hb_000776_050 |
0.1560171184 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 14 |
Hb_004032_310 |
0.156273868 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 15 |
Hb_000107_440 |
0.1569699777 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A synthase |
hydroxymethylglutaryl-CoA reductase [Hevea brasiliensis] |
| 16 |
Hb_000038_040 |
0.1584193592 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 17 |
Hb_006935_010 |
0.1587004218 |
- |
- |
PREDICTED: inositol transporter 1 isoform X1 [Gossypium raimondii] |
| 18 |
Hb_003207_100 |
0.1606060574 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP9-like [Jatropha curcas] |
| 19 |
Hb_001863_450 |
0.1613974502 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 20 |
Hb_001258_150 |
0.1617699656 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |