| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
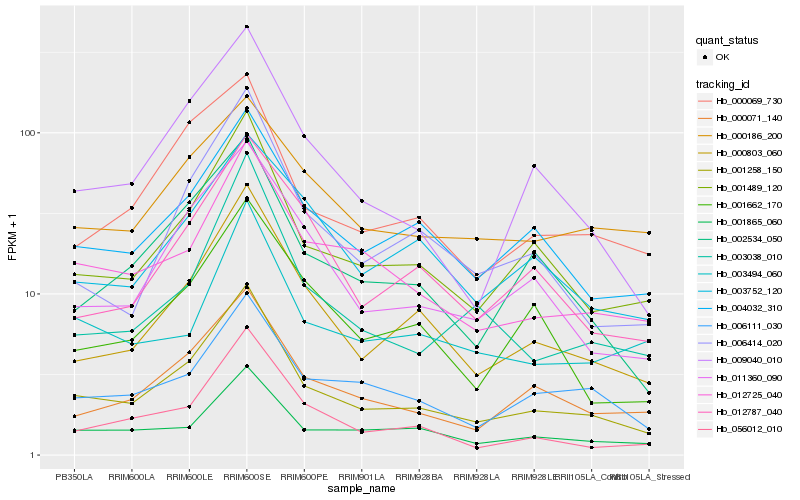
Hb_001258_150 |
0.0 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |
| 2 |
Hb_001489_120 |
0.086525137 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000803_060 |
0.1037282225 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 4 |
Hb_004032_310 |
0.1096762047 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 5 |
Hb_012725_040 |
0.1116424815 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 6 |
Hb_000071_140 |
0.1124942291 |
- |
- |
PREDICTED: uncharacterized protein LOC105628258 [Jatropha curcas] |
| 7 |
Hb_011360_090 |
0.1160759189 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 8 |
Hb_009040_010 |
0.1184437786 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_002534_050 |
0.1193199745 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |
| 10 |
Hb_000069_730 |
0.1209812902 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_001865_060 |
0.1212991277 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 12 |
Hb_056012_010 |
0.1222735209 |
- |
- |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 13 |
Hb_006414_020 |
0.1261000243 |
transcription factor |
TF Family: MYB |
- |
| 14 |
Hb_006111_030 |
0.1264133211 |
- |
- |
PREDICTED: phospholipase A(1) LCAT3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003038_010 |
0.1368519457 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 16 |
Hb_012787_040 |
0.1395249822 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001662_170 |
0.1396682307 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 18 |
Hb_000186_200 |
0.1408511793 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 19 |
Hb_003494_060 |
0.1429136308 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003752_120 |
0.1449186566 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |