| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
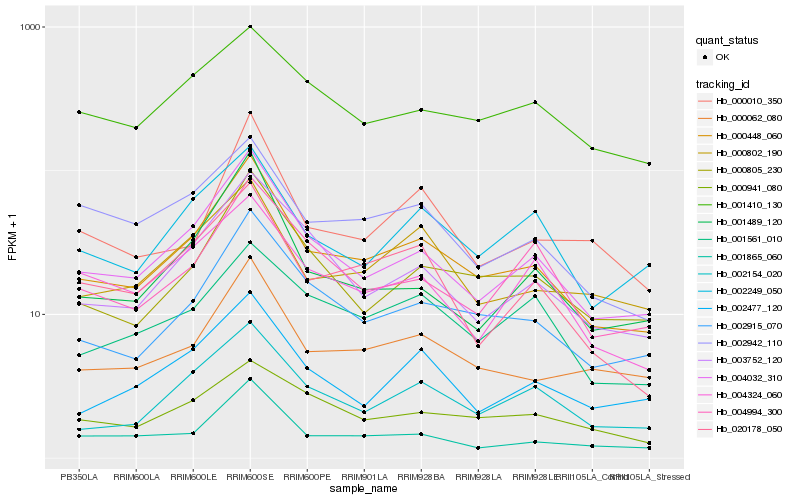
Hb_000448_060 |
0.0 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 2 |
Hb_004032_310 |
0.0748659543 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 3 |
Hb_002915_070 |
0.0855103482 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA GLABRA 1 [Jatropha curcas] |
| 4 |
Hb_000062_080 |
0.1035269842 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 5 |
Hb_002154_020 |
0.1044966789 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_001865_060 |
0.105201609 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 7 |
Hb_000805_230 |
0.1062789865 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 8 |
Hb_003752_120 |
0.1117117595 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 9 |
Hb_020178_050 |
0.1130640665 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 10 |
Hb_000802_190 |
0.1209757369 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 11 |
Hb_002249_050 |
0.1226430557 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 12 |
Hb_002942_110 |
0.1238295772 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 2-like [Jatropha curcas] |
| 13 |
Hb_001489_120 |
0.1274643028 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002477_120 |
0.1275789694 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 15 |
Hb_001561_010 |
0.1280461935 |
- |
- |
PREDICTED: uncharacterized protein LOC105635045 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004994_300 |
0.1292447633 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 17 |
Hb_000941_080 |
0.1295374617 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 7-like isoform X3 [Populus euphratica] |
| 18 |
Hb_001410_130 |
0.1311406914 |
- |
- |
hypothetical protein POPTR_0001s10880g [Populus trichocarpa] |
| 19 |
Hb_000010_350 |
0.1318498749 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 20 |
Hb_004324_060 |
0.1333988044 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |