| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
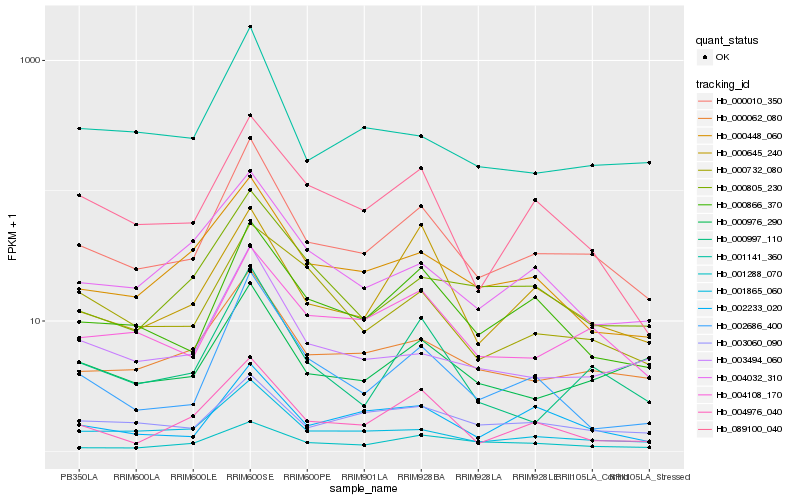
Hb_000010_350 |
0.0 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 2 |
Hb_000062_080 |
0.1067472084 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 3 |
Hb_001865_060 |
0.113577637 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 4 |
Hb_000866_370 |
0.1144485703 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 5 |
Hb_004108_170 |
0.1170690908 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000448_060 |
0.1318498749 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 7 |
Hb_089100_040 |
0.1343958393 |
- |
- |
PREDICTED: uncharacterized protein LOC105638961 [Jatropha curcas] |
| 8 |
Hb_003494_060 |
0.1378711253 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000976_290 |
0.1384847108 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105646228 [Jatropha curcas] |
| 10 |
Hb_001141_360 |
0.1389564535 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 11 |
Hb_000997_110 |
0.1393340754 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 12 |
Hb_002233_020 |
0.1402682344 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 13 |
Hb_002686_400 |
0.1450302499 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 14 |
Hb_000805_230 |
0.1476857172 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 15 |
Hb_003060_090 |
0.1493132208 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 16 |
Hb_004976_040 |
0.1494642517 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |
| 17 |
Hb_004032_310 |
0.1500919506 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 18 |
Hb_000645_240 |
0.1513399599 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 19 |
Hb_001288_070 |
0.1516652509 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 20 |
Hb_000732_080 |
0.1518464579 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g45440 [Jatropha curcas] |