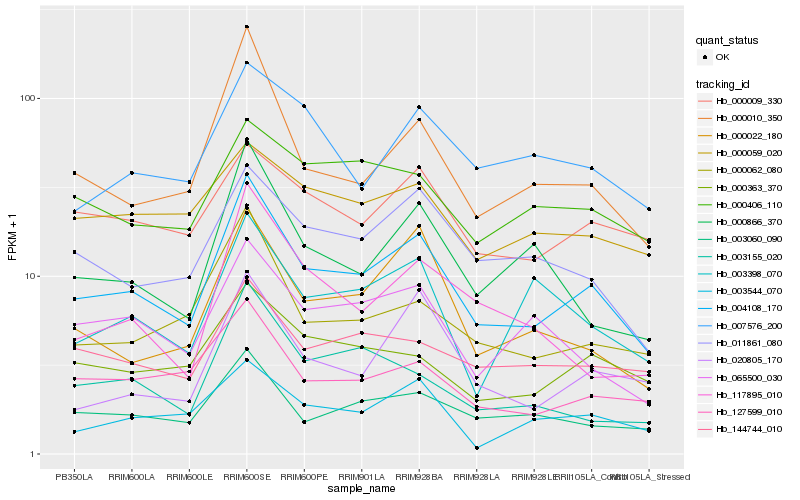
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004108_170 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000010_350 |
0.1170690908 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 3 |
Hb_003060_090 |
0.1241049249 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 4 |
Hb_000062_080 |
0.1266219827 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 5 |
Hb_127599_010 |
0.1271812364 |
- |
- |
hypothetical protein CISIN_1g0010462mg, partial [Citrus sinensis] |
| 6 |
Hb_011861_080 |
0.1310209756 |
- |
- |
hypothetical protein JCGZ_06088 [Jatropha curcas] |
| 7 |
Hb_000363_370 |
0.1377438609 |
- |
- |
PREDICTED: putative BPI/LBP family protein At1g04970 [Jatropha curcas] |
| 8 |
Hb_065500_030 |
0.137789938 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 [Jatropha curcas] |
| 9 |
Hb_000866_370 |
0.1383960306 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 10 |
Hb_000022_180 |
0.1389266133 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_144744_010 |
0.1404758007 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 12 |
Hb_000009_330 |
0.1436343725 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 13 |
Hb_003544_070 |
0.1457548193 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_007576_200 |
0.1497179023 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 15 |
Hb_020805_170 |
0.1513460663 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 16 |
Hb_003398_070 |
0.1513899188 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 17 |
Hb_003155_020 |
0.1523732736 |
- |
- |
Rpp4C3 [Medicago truncatula] |
| 18 |
Hb_000406_110 |
0.1525289232 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1 [Jatropha curcas] |
| 19 |
Hb_117895_010 |
0.1545936383 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 20 |
Hb_000059_020 |
0.1546344186 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |