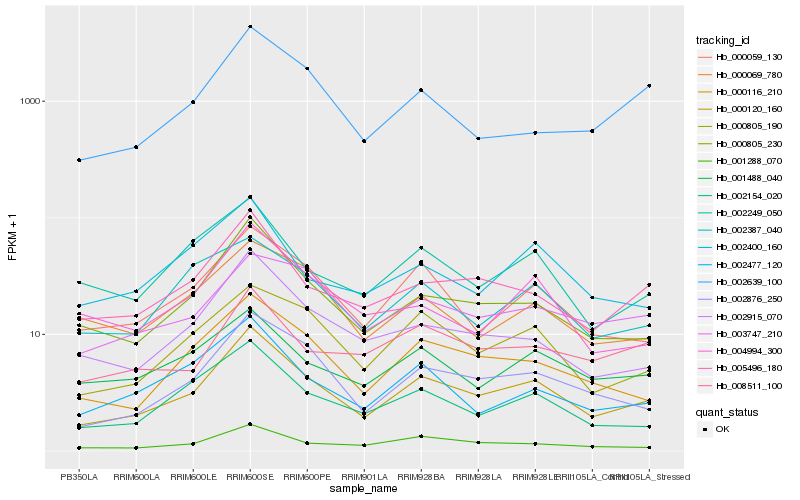
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_160 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_000805_230 |
0.1020205287 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 3 |
Hb_002876_250 |
0.10513437 |
- |
- |
PREDICTED: receptor-like protein kinase 2 [Jatropha curcas] |
| 4 |
Hb_000116_210 |
0.1131943686 |
- |
- |
PREDICTED: uncharacterized protein LOC105628567 [Jatropha curcas] |
| 5 |
Hb_005496_180 |
0.1162924905 |
- |
- |
PREDICTED: U-box domain-containing protein 4-like [Jatropha curcas] |
| 6 |
Hb_001288_070 |
0.1181858144 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 7 |
Hb_002154_020 |
0.1195931748 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_000059_130 |
0.1200699795 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 9 |
Hb_000805_190 |
0.1207056739 |
transcription factor |
TF Family: GRAS |
PREDICTED: uncharacterized protein LOC103323607 [Prunus mume] |
| 10 |
Hb_002915_070 |
0.1236648196 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA GLABRA 1 [Jatropha curcas] |
| 11 |
Hb_001488_040 |
0.1367818443 |
- |
- |
PREDICTED: nipped-B-like protein A [Jatropha curcas] |
| 12 |
Hb_002477_120 |
0.1368047911 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 13 |
Hb_002249_050 |
0.1385036286 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 14 |
Hb_002387_040 |
0.1421468257 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 15 |
Hb_003747_210 |
0.1428721845 |
- |
- |
PREDICTED: uncharacterized protein LOC105630240 [Jatropha curcas] |
| 16 |
Hb_000069_780 |
0.1439148713 |
- |
- |
atpob1, putative [Ricinus communis] |
| 17 |
Hb_002400_160 |
0.1441102274 |
- |
- |
Ubiquitin-protein ligase BRE1A, putative [Ricinus communis] |
| 18 |
Hb_008511_100 |
0.1447790642 |
- |
- |
hypothetical protein POPTR_0009s11860g [Populus trichocarpa] |
| 19 |
Hb_004994_300 |
0.1449672882 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 20 |
Hb_002639_100 |
0.1451515947 |
- |
- |
dehydrin [Hevea brasiliensis] |