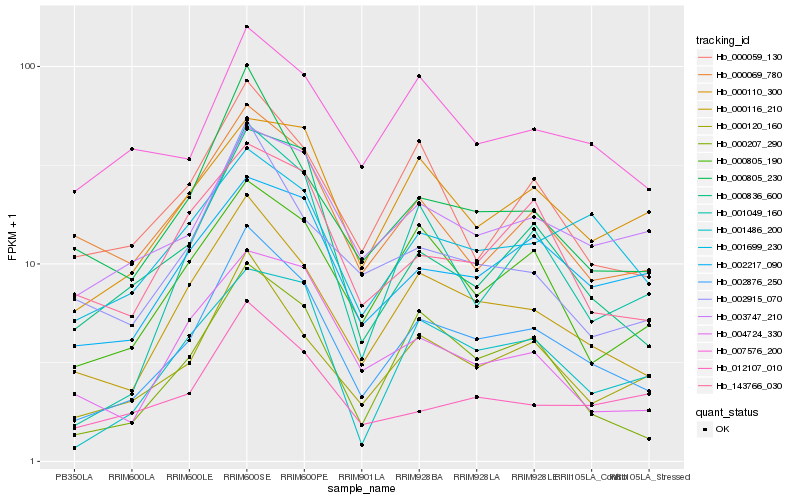
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002876_250 |
0.0 |
- |
- |
PREDICTED: receptor-like protein kinase 2 [Jatropha curcas] |
| 2 |
Hb_000116_210 |
0.085169584 |
- |
- |
PREDICTED: uncharacterized protein LOC105628567 [Jatropha curcas] |
| 3 |
Hb_000120_160 |
0.10513437 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_000207_290 |
0.1216091193 |
- |
- |
hypothetical protein JCGZ_03920 [Jatropha curcas] |
| 5 |
Hb_000805_230 |
0.1221077003 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 6 |
Hb_000836_600 |
0.1268499371 |
- |
- |
PREDICTED: delta(8)-fatty-acid desaturase 2 [Jatropha curcas] |
| 7 |
Hb_000805_190 |
0.1292519357 |
transcription factor |
TF Family: GRAS |
PREDICTED: uncharacterized protein LOC103323607 [Prunus mume] |
| 8 |
Hb_001049_160 |
0.1308759076 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 9 |
Hb_001486_200 |
0.1373631735 |
- |
- |
PREDICTED: uncharacterized protein LOC105632606 [Jatropha curcas] |
| 10 |
Hb_000059_130 |
0.1374618124 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 11 |
Hb_002217_090 |
0.1395823441 |
- |
- |
PREDICTED: probable protein phosphatase 2C 13 [Jatropha curcas] |
| 12 |
Hb_003747_210 |
0.1396774745 |
- |
- |
PREDICTED: uncharacterized protein LOC105630240 [Jatropha curcas] |
| 13 |
Hb_002915_070 |
0.1424161544 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA GLABRA 1 [Jatropha curcas] |
| 14 |
Hb_143766_030 |
0.1447467268 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_012107_010 |
0.145222339 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 16 |
Hb_001699_230 |
0.1453127414 |
- |
- |
PREDICTED: calcium-dependent protein kinase 28 [Jatropha curcas] |
| 17 |
Hb_000110_300 |
0.1472698811 |
- |
- |
PREDICTED: uncharacterized protein LOC105642013 [Jatropha curcas] |
| 18 |
Hb_004724_330 |
0.1480506313 |
- |
- |
Capsanthin/capsorubin synthase, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_000069_780 |
0.1490575393 |
- |
- |
atpob1, putative [Ricinus communis] |
| 20 |
Hb_007576_200 |
0.1491052583 |
- |
- |
amino acid transporter, putative [Ricinus communis] |