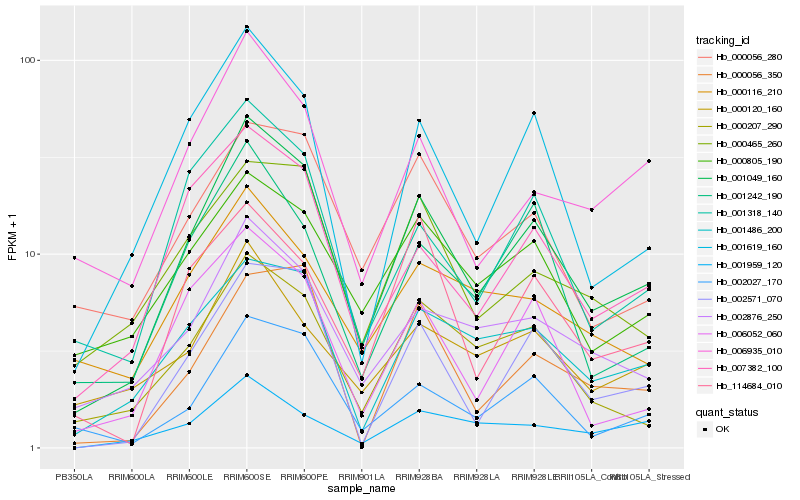
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001049_160 |
0.0 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 2 |
Hb_001619_160 |
0.1248988724 |
transcription factor |
TF Family: ERF |
DREB1p [Hevea brasiliensis] |
| 3 |
Hb_002027_170 |
0.128067659 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RKF3 [Jatropha curcas] |
| 4 |
Hb_007382_100 |
0.1298877143 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 5 |
Hb_002876_250 |
0.1308759076 |
- |
- |
PREDICTED: receptor-like protein kinase 2 [Jatropha curcas] |
| 6 |
Hb_001486_200 |
0.1349033283 |
- |
- |
PREDICTED: uncharacterized protein LOC105632606 [Jatropha curcas] |
| 7 |
Hb_001318_140 |
0.1362751408 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 8 |
Hb_000207_290 |
0.1407748081 |
- |
- |
hypothetical protein JCGZ_03920 [Jatropha curcas] |
| 9 |
Hb_002571_070 |
0.1417532956 |
- |
- |
PREDICTED: cation/calcium exchanger 2 [Jatropha curcas] |
| 10 |
Hb_006052_060 |
0.1469513694 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 11 |
Hb_006935_010 |
0.1478926708 |
- |
- |
PREDICTED: inositol transporter 1 isoform X1 [Gossypium raimondii] |
| 12 |
Hb_001242_190 |
0.1500575492 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 1 isoform X3 [Jatropha curcas] |
| 13 |
Hb_000056_280 |
0.1522929358 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 14 |
Hb_000805_190 |
0.1526834331 |
transcription factor |
TF Family: GRAS |
PREDICTED: uncharacterized protein LOC103323607 [Prunus mume] |
| 15 |
Hb_000465_260 |
0.1528779674 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT4-like [Jatropha curcas] |
| 16 |
Hb_000120_160 |
0.1630348638 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_114684_010 |
0.1664451222 |
- |
- |
PREDICTED: seed maturation protein PM36 [Jatropha curcas] |
| 18 |
Hb_001959_120 |
0.1669063888 |
- |
- |
PREDICTED: uncharacterized protein LOC105638487 [Jatropha curcas] |
| 19 |
Hb_000116_210 |
0.1679806043 |
- |
- |
PREDICTED: uncharacterized protein LOC105628567 [Jatropha curcas] |
| 20 |
Hb_000056_350 |
0.1681640609 |
- |
- |
PREDICTED: uncharacterized protein LOC105630437 [Jatropha curcas] |