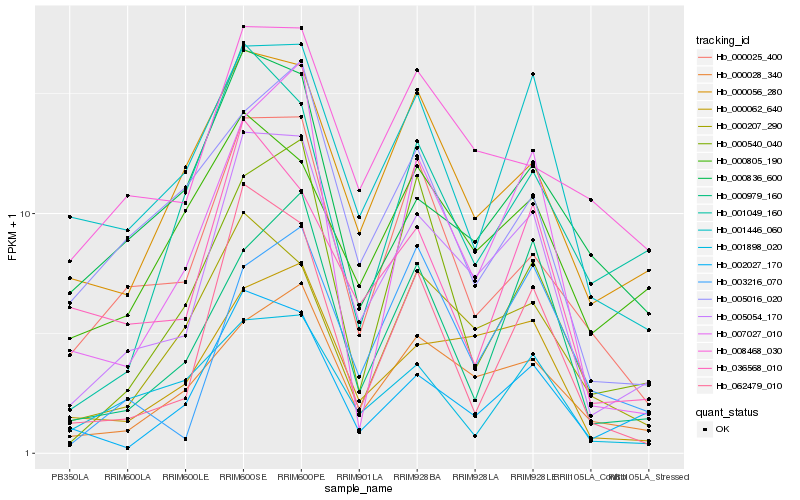
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005054_170 |
0.0 |
transcription factor |
TF Family: MYB |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_000062_640 |
0.1320307918 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF2.2-like isoform X2 [Populus euphratica] |
| 3 |
Hb_000028_340 |
0.1457532799 |
- |
- |
PREDICTED: potassium channel SKOR-like [Prunus mume] |
| 4 |
Hb_002027_170 |
0.1526410155 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RKF3 [Jatropha curcas] |
| 5 |
Hb_062479_010 |
0.1588082546 |
- |
- |
hypothetical protein PHAVU_008G071200g [Phaseolus vulgaris] |
| 6 |
Hb_000056_280 |
0.1603348567 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 7 |
Hb_000025_400 |
0.1635326626 |
- |
- |
PREDICTED: uncharacterized protein LOC105110770 [Populus euphratica] |
| 8 |
Hb_001446_060 |
0.1675676552 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_000207_290 |
0.1680890051 |
- |
- |
hypothetical protein JCGZ_03920 [Jatropha curcas] |
| 10 |
Hb_000979_160 |
0.1720542228 |
- |
- |
hypothetical protein RCOM_0629030 [Ricinus communis] |
| 11 |
Hb_007027_010 |
0.1749501345 |
- |
- |
RING-H2 finger protein ATL2J, putative [Ricinus communis] |
| 12 |
Hb_008468_030 |
0.1780813105 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 13 |
Hb_000836_600 |
0.1833955554 |
- |
- |
PREDICTED: delta(8)-fatty-acid desaturase 2 [Jatropha curcas] |
| 14 |
Hb_036568_010 |
0.1836991794 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 23 [Jatropha curcas] |
| 15 |
Hb_000540_040 |
0.1858838828 |
- |
- |
PREDICTED: chitinase 2 [Amborella trichopoda] |
| 16 |
Hb_001898_020 |
0.1885070049 |
- |
- |
PREDICTED: uncharacterized protein LOC105629659 [Jatropha curcas] |
| 17 |
Hb_005016_020 |
0.1894468518 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_001049_160 |
0.1939130545 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 19 |
Hb_003216_070 |
0.1947672151 |
- |
- |
hypothetical protein RCOM_0483300 [Ricinus communis] |
| 20 |
Hb_000805_190 |
0.1952246625 |
transcription factor |
TF Family: GRAS |
PREDICTED: uncharacterized protein LOC103323607 [Prunus mume] |