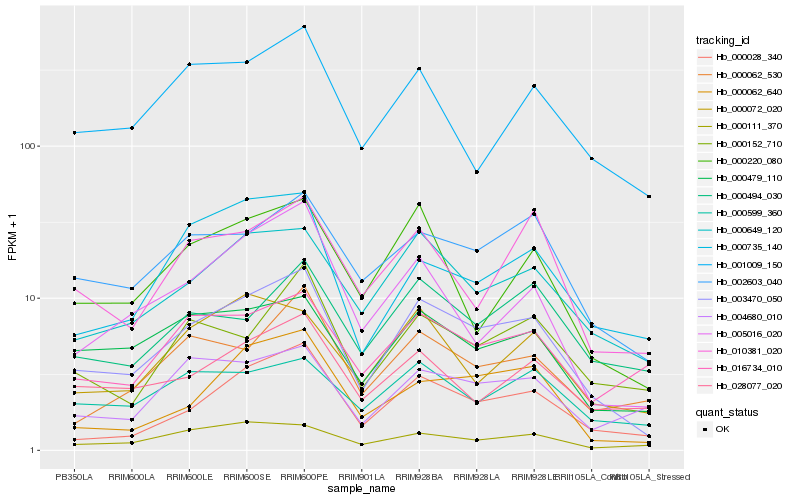
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003470_050 |
0.0 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_000649_120 |
0.1217351011 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_002603_040 |
0.1251540857 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 2-like [Jatropha curcas] |
| 4 |
Hb_000028_340 |
0.1313903111 |
- |
- |
PREDICTED: potassium channel SKOR-like [Prunus mume] |
| 5 |
Hb_000479_110 |
0.1354263401 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 8, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000062_640 |
0.1459705359 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF2.2-like isoform X2 [Populus euphratica] |
| 7 |
Hb_000220_080 |
0.1487256988 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_000152_710 |
0.1527614347 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 1 [Jatropha curcas] |
| 9 |
Hb_000111_370 |
0.15324916 |
- |
- |
PREDICTED: cation/H(+) antiporter 2-like [Jatropha curcas] |
| 10 |
Hb_005016_020 |
0.1553343036 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_000599_360 |
0.1554941183 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 12 |
Hb_010381_020 |
0.1562963873 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 13 |
Hb_001009_150 |
0.1563690314 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase [Gossypium arboreum] |
| 14 |
Hb_028077_020 |
0.1566303787 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000735_140 |
0.1579765509 |
- |
- |
3-glucanase family protein [Populus trichocarpa] |
| 16 |
Hb_000072_020 |
0.1581488482 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000494_030 |
0.1594178462 |
- |
- |
PREDICTED: uncharacterized protein LOC105643196 [Jatropha curcas] |
| 18 |
Hb_000062_530 |
0.159631631 |
- |
- |
PREDICTED: GDP-mannose transporter GONST3-like [Populus euphratica] |
| 19 |
Hb_004680_010 |
0.1598838864 |
- |
- |
PREDICTED: uncharacterized protein LOC105636246 isoform X1 [Jatropha curcas] |
| 20 |
Hb_016734_010 |
0.1609468929 |
- |
- |
PREDICTED: probable DNA-3-methyladenine glycosylase 2 [Jatropha curcas] |