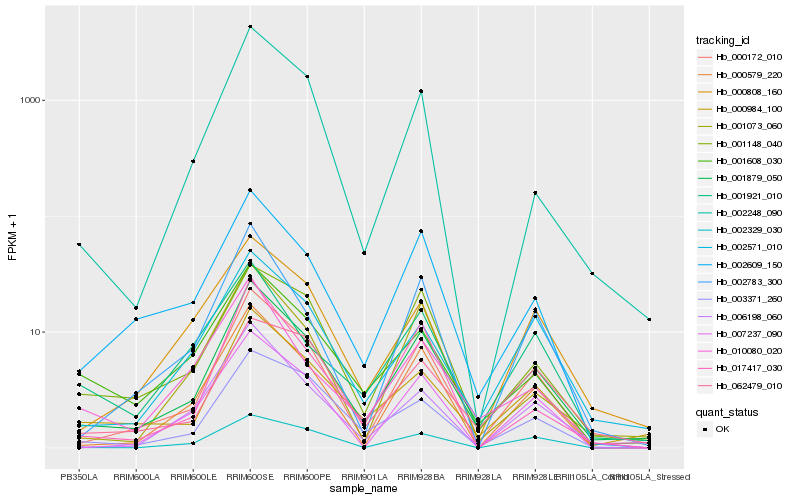
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001879_050 |
0.0 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 2 |
Hb_000172_010 |
0.1003418523 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670-like isoform X1 [Populus euphratica] |
| 3 |
Hb_000984_100 |
0.1040798262 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 4 |
Hb_002609_150 |
0.1194577355 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002571_010 |
0.126539158 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 6 |
Hb_001148_040 |
0.1334714791 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 7 |
Hb_000579_220 |
0.1335786017 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_002783_300 |
0.1435567331 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_003371_260 |
0.1437682174 |
- |
- |
hypothetical protein EUGRSUZ_B03661 [Eucalyptus grandis] |
| 10 |
Hb_002329_030 |
0.1444904463 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 2-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000808_160 |
0.1452289329 |
- |
- |
PREDICTED: uncharacterized protein LOC105647443 [Jatropha curcas] |
| 12 |
Hb_001073_060 |
0.1492536943 |
- |
- |
phosphate transporter 1 [Hevea brasiliensis] |
| 13 |
Hb_006198_060 |
0.1520392069 |
- |
- |
PREDICTED: protein SULFUR DEFICIENCY-INDUCED 1 [Jatropha curcas] |
| 14 |
Hb_001608_030 |
0.1527584361 |
- |
- |
Protein NSP-INTERACTING KINASE 1 [Glycine soja] |
| 15 |
Hb_010080_020 |
0.1527723534 |
- |
- |
PREDICTED: uncharacterized protein LOC105635249 [Jatropha curcas] |
| 16 |
Hb_002248_090 |
0.1532080159 |
- |
- |
metallothionein 3-like protein [Hevea brasiliensis] |
| 17 |
Hb_001921_010 |
0.15518316 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 18 |
Hb_062479_010 |
0.1552092328 |
- |
- |
hypothetical protein PHAVU_008G071200g [Phaseolus vulgaris] |
| 19 |
Hb_007237_090 |
0.1554447188 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_017417_030 |
0.1557635285 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |