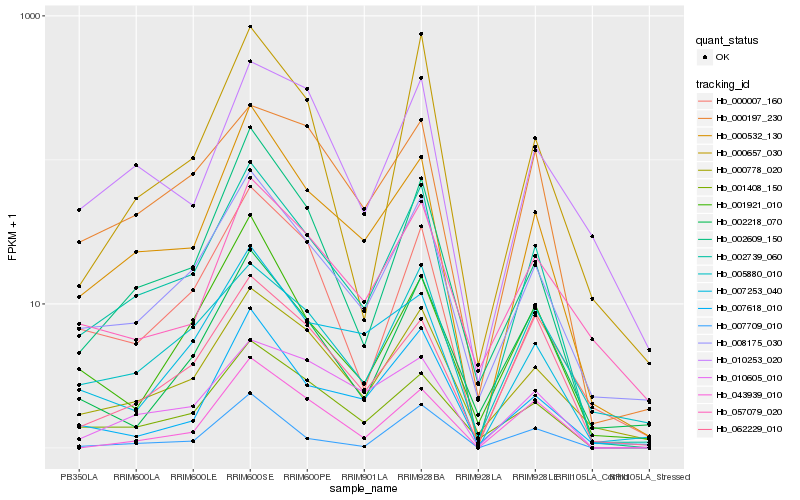
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002739_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_008175_030 |
0.0895330748 |
transcription factor |
TF Family: TAZ |
PREDICTED: BTB/POZ and TAZ domain-containing protein 3 [Jatropha curcas] |
| 3 |
Hb_000532_130 |
0.0959497278 |
- |
- |
hypothetical protein [Anabaena sp. PCC 7108] |
| 4 |
Hb_000778_020 |
0.1168627542 |
- |
- |
PREDICTED: salicylic acid-binding protein 2-like [Jatropha curcas] |
| 5 |
Hb_001408_150 |
0.1205694006 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005880_010 |
0.1361706122 |
- |
- |
PREDICTED: uncharacterized protein LOC105646431 [Jatropha curcas] |
| 7 |
Hb_002609_150 |
0.1364549671 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000007_160 |
0.1471881529 |
- |
- |
hypothetical protein POPTR_0008s10970g [Populus trichocarpa] |
| 9 |
Hb_000197_230 |
0.1472939981 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 10 |
Hb_007618_010 |
0.1484787077 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 11 |
Hb_062229_010 |
0.1492764574 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 12 |
Hb_007253_040 |
0.1539829095 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-10 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000657_030 |
0.1544344251 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 14 |
Hb_002218_070 |
0.1575316735 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 15 |
Hb_057079_020 |
0.1584005761 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_010605_010 |
0.1590248015 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 17 |
Hb_007709_010 |
0.1607464866 |
- |
- |
- |
| 18 |
Hb_010253_020 |
0.1616222387 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_001921_010 |
0.1623306628 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 20 |
Hb_043939_010 |
0.1642175697 |
- |
- |
hypothetical protein B456_N011100 [Gossypium raimondii] |