| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
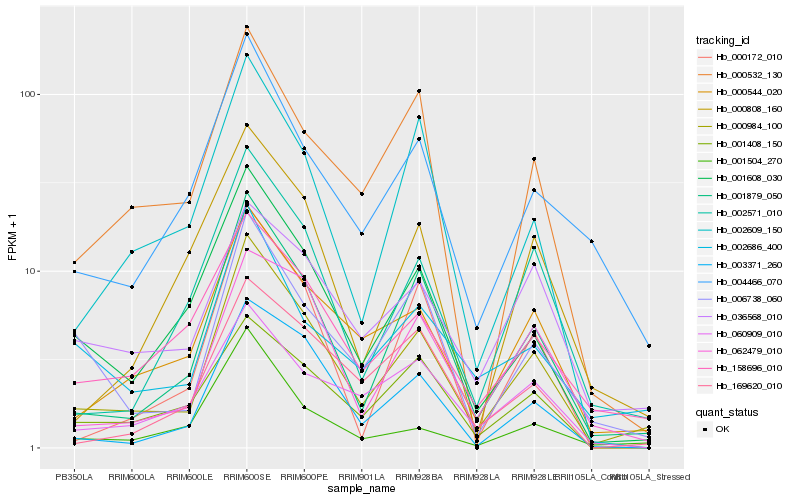
Hb_000984_100 |
0.0 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 2 |
Hb_001879_050 |
0.1040798262 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 3 |
Hb_000544_020 |
0.1286701755 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 4 |
Hb_001608_030 |
0.1439115976 |
- |
- |
Protein NSP-INTERACTING KINASE 1 [Glycine soja] |
| 5 |
Hb_002571_010 |
0.1460827581 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 6 |
Hb_003371_260 |
0.1502716233 |
- |
- |
hypothetical protein EUGRSUZ_B03661 [Eucalyptus grandis] |
| 7 |
Hb_004466_070 |
0.1593959351 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 8 |
Hb_006738_060 |
0.159691847 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_002686_400 |
0.1603798423 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 10 |
Hb_158696_010 |
0.1604903119 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 11 |
Hb_062479_010 |
0.1614031484 |
- |
- |
hypothetical protein PHAVU_008G071200g [Phaseolus vulgaris] |
| 12 |
Hb_000172_010 |
0.1642167511 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670-like isoform X1 [Populus euphratica] |
| 13 |
Hb_002609_150 |
0.1660433744 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000532_130 |
0.1661959951 |
- |
- |
hypothetical protein [Anabaena sp. PCC 7108] |
| 15 |
Hb_001504_270 |
0.1695502748 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 16 |
Hb_001408_150 |
0.1725126332 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000808_160 |
0.1742290995 |
- |
- |
PREDICTED: uncharacterized protein LOC105647443 [Jatropha curcas] |
| 18 |
Hb_036568_010 |
0.1746647181 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 23 [Jatropha curcas] |
| 19 |
Hb_169620_010 |
0.17522142 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 20 |
Hb_060909_010 |
0.1756925388 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |