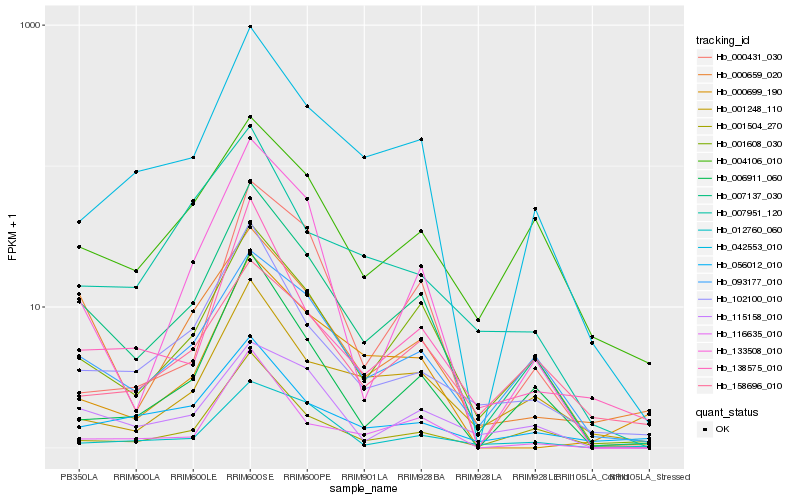
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007137_030 |
0.0 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.3-like [Jatropha curcas] |
| 2 |
Hb_001608_030 |
0.1048124221 |
- |
- |
Protein NSP-INTERACTING KINASE 1 [Glycine soja] |
| 3 |
Hb_093177_010 |
0.1273274256 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 4 |
Hb_042553_010 |
0.1383438801 |
- |
- |
PREDICTED: glutathione S-transferase T1-like [Jatropha curcas] |
| 5 |
Hb_133508_010 |
0.1588010809 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.3-like [Jatropha curcas] |
| 6 |
Hb_102100_010 |
0.161369414 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 7 |
Hb_115158_010 |
0.1615007852 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 8 |
Hb_006911_060 |
0.1655157667 |
- |
- |
hypothetical protein CISIN_1g000594mg [Citrus sinensis] |
| 9 |
Hb_000659_020 |
0.1671858876 |
- |
- |
PREDICTED: putative L-type lectin-domain containing receptor kinase II.2 [Jatropha curcas] |
| 10 |
Hb_004106_010 |
0.1753065733 |
- |
- |
PREDICTED: protein GPR107 isoform X1 [Jatropha curcas] |
| 11 |
Hb_116635_010 |
0.179586886 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 12 |
Hb_138575_010 |
0.1797627151 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 13 |
Hb_001504_270 |
0.1822674193 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 14 |
Hb_001248_110 |
0.1826886132 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 15 |
Hb_056012_010 |
0.1853659664 |
- |
- |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 16 |
Hb_158696_010 |
0.189023417 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 17 |
Hb_012760_060 |
0.1929043176 |
- |
- |
PREDICTED: uncharacterized protein LOC105647085 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007951_120 |
0.1930357268 |
- |
- |
PREDICTED: uncharacterized protein At1g15400 [Populus euphratica] |
| 19 |
Hb_000699_190 |
0.194545342 |
- |
- |
unknown [Populus trichocarpa] |
| 20 |
Hb_000431_030 |
0.1986557058 |
transcription factor |
TF Family: C2H2 |
zinc finger family protein [Populus trichocarpa] |