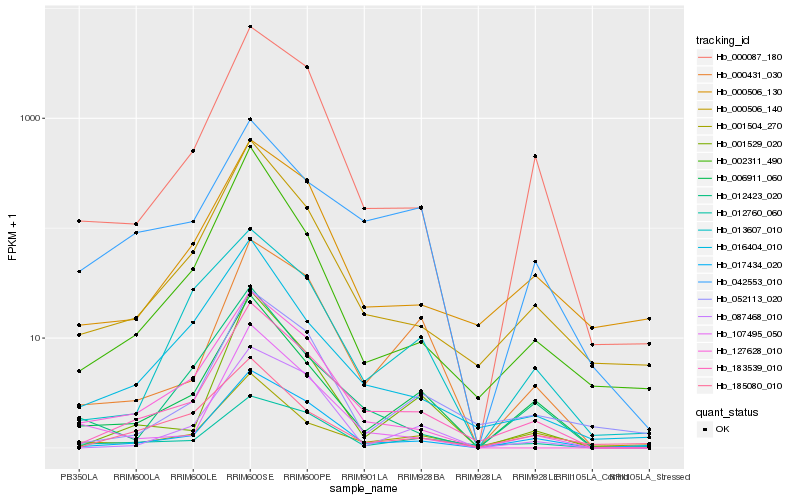
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_183539_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0005s04441g [Populus trichocarpa] |
| 2 |
Hb_017434_020 |
0.1180294299 |
transcription factor |
TF Family: C2H2 |
Zinc finger protein 4 [Populus trichocarpa] |
| 3 |
Hb_000506_140 |
0.130202821 |
- |
- |
hypothetical protein PRUPE_ppb019425mg [Prunus persica] |
| 4 |
Hb_000087_180 |
0.141028374 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_006911_060 |
0.1471060492 |
- |
- |
hypothetical protein CISIN_1g000594mg [Citrus sinensis] |
| 6 |
Hb_000506_130 |
0.1577125935 |
- |
- |
DGCR14-related family protein [Populus trichocarpa] |
| 7 |
Hb_001529_020 |
0.1625632885 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_052113_020 |
0.164208447 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 9 |
Hb_016404_010 |
0.1656464491 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 10 |
Hb_127628_010 |
0.1674336119 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 11 |
Hb_042553_010 |
0.1686367649 |
- |
- |
PREDICTED: glutathione S-transferase T1-like [Jatropha curcas] |
| 12 |
Hb_000431_030 |
0.1697697369 |
transcription factor |
TF Family: C2H2 |
zinc finger family protein [Populus trichocarpa] |
| 13 |
Hb_012760_060 |
0.1719604254 |
- |
- |
PREDICTED: uncharacterized protein LOC105647085 isoform X1 [Jatropha curcas] |
| 14 |
Hb_107495_050 |
0.1732577539 |
transcription factor |
TF Family: MIKC |
mads box protein, putative [Ricinus communis] |
| 15 |
Hb_002311_490 |
0.1749707977 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 16 |
Hb_001504_270 |
0.1764552979 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 17 |
Hb_087468_010 |
0.179524419 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 18 |
Hb_185080_010 |
0.1798262563 |
- |
- |
- |
| 19 |
Hb_012423_020 |
0.1806963055 |
- |
- |
hypothetical protein JCGZ_20730 [Jatropha curcas] |
| 20 |
Hb_013607_010 |
0.1807487607 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA3 [Jatropha curcas] |