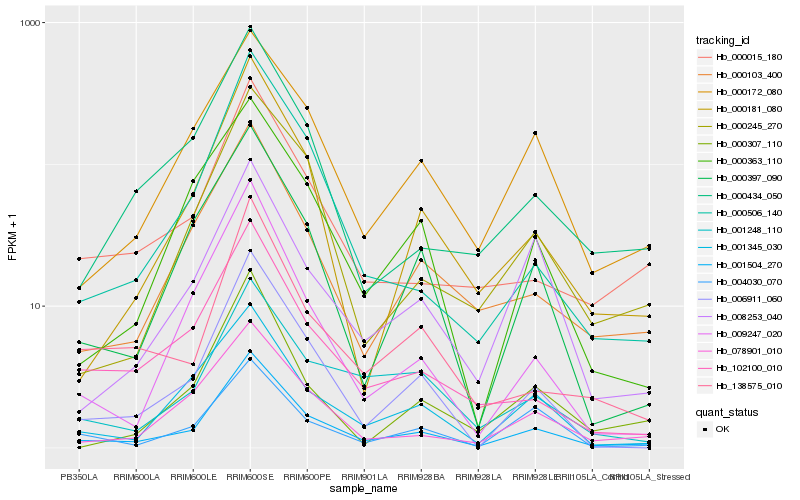
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001504_270 |
0.0 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 2 |
Hb_006911_060 |
0.1156934816 |
- |
- |
hypothetical protein CISIN_1g000594mg [Citrus sinensis] |
| 3 |
Hb_000181_080 |
0.1379082149 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 4 |
Hb_000172_080 |
0.1385179132 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 5 |
Hb_001345_030 |
0.1393697689 |
- |
- |
hypothetical protein POPTR_0007s10390g, partial [Populus trichocarpa] |
| 6 |
Hb_000397_090 |
0.140904505 |
- |
- |
PREDICTED: uncharacterized protein LOC105641235 [Jatropha curcas] |
| 7 |
Hb_078901_010 |
0.1421638247 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 8 |
Hb_000103_400 |
0.1423934801 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000015_180 |
0.1479713015 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 10 |
Hb_102100_010 |
0.1488466046 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 11 |
Hb_009247_020 |
0.1491540724 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 12 |
Hb_000506_140 |
0.1501130143 |
- |
- |
hypothetical protein PRUPE_ppb019425mg [Prunus persica] |
| 13 |
Hb_000245_270 |
0.1509619555 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 14 |
Hb_000434_050 |
0.1563767119 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 15 |
Hb_138575_010 |
0.1565683249 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 16 |
Hb_001248_110 |
0.1570284405 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 17 |
Hb_008253_040 |
0.1571627258 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 18 |
Hb_000363_110 |
0.158444867 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 21 [Jatropha curcas] |
| 19 |
Hb_000307_110 |
0.1590149349 |
- |
- |
hypothetical protein JCGZ_08160 [Jatropha curcas] |
| 20 |
Hb_004030_070 |
0.1612865355 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |