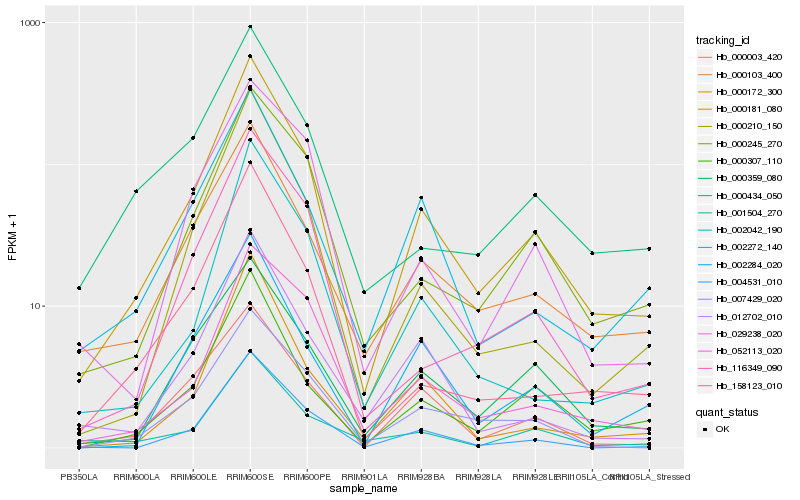
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000181_080 |
0.0 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 2 |
Hb_000307_110 |
0.113220786 |
- |
- |
hypothetical protein JCGZ_08160 [Jatropha curcas] |
| 3 |
Hb_004531_010 |
0.116745464 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At2g01680-like [Jatropha curcas] |
| 4 |
Hb_000103_400 |
0.1167862878 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000245_270 |
0.1197993155 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 6 |
Hb_000210_150 |
0.1246054393 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 7 |
Hb_007429_020 |
0.1287701825 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_000003_420 |
0.1295522982 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000172_300 |
0.130904678 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 10 |
Hb_002284_020 |
0.1325162629 |
- |
- |
PREDICTED: probable inorganic phosphate transporter 1-9 [Jatropha curcas] |
| 11 |
Hb_012702_010 |
0.1327884514 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X4 [Vitis vinifera] |
| 12 |
Hb_002272_140 |
0.1345378955 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL6-like [Jatropha curcas] |
| 13 |
Hb_000359_080 |
0.13526411 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 14 |
Hb_001504_270 |
0.1379082149 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 15 |
Hb_116349_090 |
0.1380331176 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 9 [Jatropha curcas] |
| 16 |
Hb_158123_010 |
0.1382302654 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 17 |
Hb_052113_020 |
0.1413446563 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 18 |
Hb_002042_190 |
0.142621834 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL6-like [Jatropha curcas] |
| 19 |
Hb_029238_020 |
0.1428421507 |
- |
- |
PREDICTED: expansin-like A1 [Jatropha curcas] |
| 20 |
Hb_000434_050 |
0.1448422936 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |