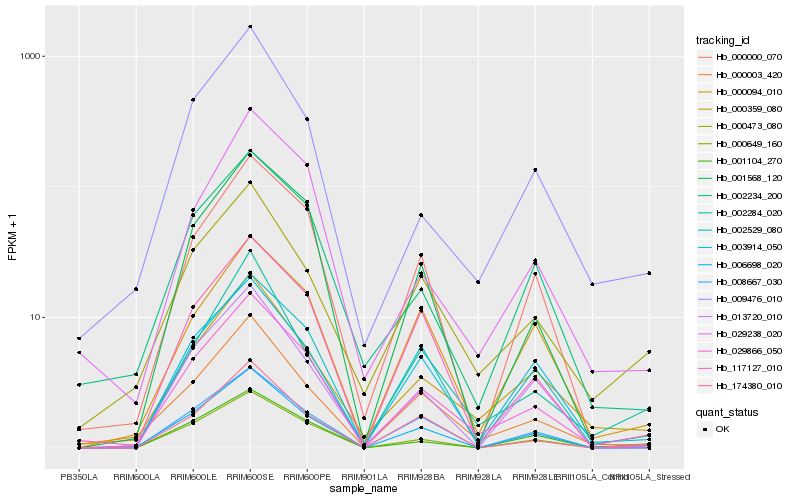
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029866_050 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF4 [Jatropha curcas] |
| 2 |
Hb_006698_020 |
0.1020261642 |
- |
- |
hypothetical protein JCGZ_04585 [Jatropha curcas] |
| 3 |
Hb_000649_160 |
0.1077585813 |
- |
- |
PREDICTED: nudix hydrolase 18, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000003_420 |
0.1112970144 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 5 |
Hb_001568_120 |
0.113946992 |
- |
- |
hypothetical protein JCGZ_05918 [Jatropha curcas] |
| 6 |
Hb_000359_080 |
0.1197348181 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 7 |
Hb_000473_080 |
0.1264676409 |
- |
- |
PREDICTED: lipase-like PAD4 [Populus euphratica] |
| 8 |
Hb_003914_050 |
0.1282367091 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 9 |
Hb_000000_070 |
0.1290195386 |
- |
- |
allene oxide synthase [Hevea brasiliensis] |
| 10 |
Hb_174380_010 |
0.1291888732 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_117127_010 |
0.1295294662 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH106 [Jatropha curcas] |
| 12 |
Hb_002529_080 |
0.1339926535 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase [Jatropha curcas] |
| 13 |
Hb_002284_020 |
0.1347388755 |
- |
- |
PREDICTED: probable inorganic phosphate transporter 1-9 [Jatropha curcas] |
| 14 |
Hb_009476_010 |
0.1364736117 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 15 |
Hb_008667_030 |
0.1372488167 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 16 |
Hb_002234_200 |
0.137692642 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |
| 17 |
Hb_029238_020 |
0.139187428 |
- |
- |
PREDICTED: expansin-like A1 [Jatropha curcas] |
| 18 |
Hb_000094_010 |
0.1394198326 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 19 |
Hb_013720_010 |
0.1427817576 |
- |
- |
Putative DNA repair RAD23-3 -like protein [Gossypium arboreum] |
| 20 |
Hb_001104_270 |
0.1466206412 |
- |
- |
hypothetical protein POPTR_0007s13980g [Populus trichocarpa] |