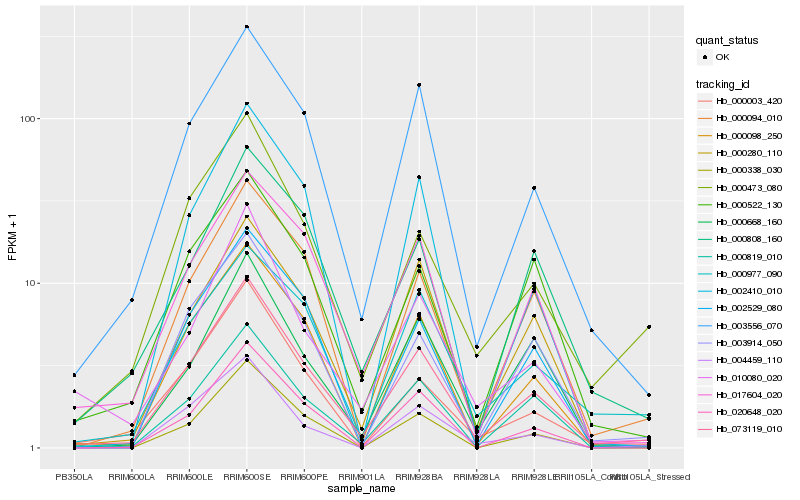
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_073119_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 2 |
Hb_003556_070 |
0.0841477353 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 3 |
Hb_000098_250 |
0.108398032 |
- |
- |
UDP-glucose flavonoid 3-O-glucosyltransferase [Medicago truncatula] |
| 4 |
Hb_000819_010 |
0.1088331525 |
- |
- |
Receptor-like protein kinase 1, putative [Theobroma cacao] |
| 5 |
Hb_000094_010 |
0.1112619969 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 6 |
Hb_017604_020 |
0.1154410096 |
- |
- |
hypothetical protein JCGZ_22673 [Jatropha curcas] |
| 7 |
Hb_000003_420 |
0.1158040208 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_000473_080 |
0.1197111942 |
- |
- |
PREDICTED: lipase-like PAD4 [Populus euphratica] |
| 9 |
Hb_000280_110 |
0.122472443 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 10 |
Hb_020648_020 |
0.122556544 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor pho1, putative [Ricinus communis] |
| 11 |
Hb_003914_050 |
0.123318737 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 12 |
Hb_000522_130 |
0.1250089454 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 13 |
Hb_010080_020 |
0.129247829 |
- |
- |
PREDICTED: uncharacterized protein LOC105635249 [Jatropha curcas] |
| 14 |
Hb_002529_080 |
0.1310440491 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase [Jatropha curcas] |
| 15 |
Hb_000977_090 |
0.1313134002 |
- |
- |
hypothetical protein POPTR_0010s18550g [Populus trichocarpa] |
| 16 |
Hb_000808_160 |
0.1331122106 |
- |
- |
PREDICTED: uncharacterized protein LOC105647443 [Jatropha curcas] |
| 17 |
Hb_004459_110 |
0.1339295395 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X2 [Populus euphratica] |
| 18 |
Hb_002410_010 |
0.134179447 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 19 |
Hb_000338_030 |
0.1346926922 |
transcription factor |
TF Family: HB |
hypothetical protein JCGZ_21254 [Jatropha curcas] |
| 20 |
Hb_000668_160 |
0.1348524521 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |