| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
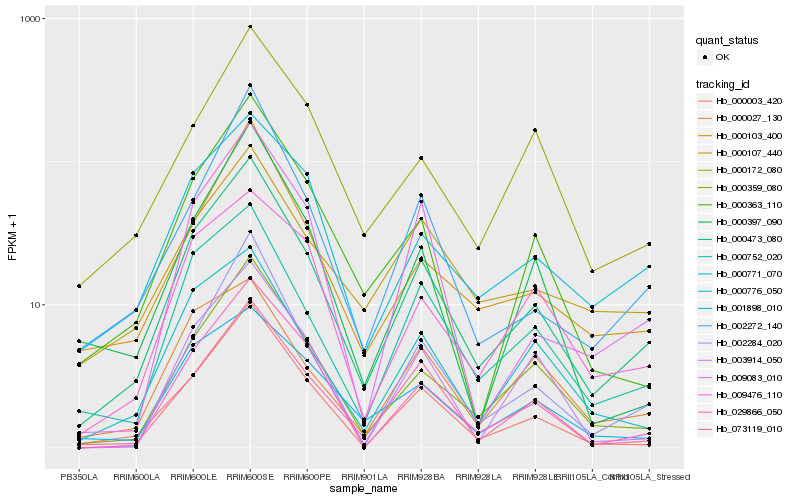
Hb_000473_080 |
0.0 |
- |
- |
PREDICTED: lipase-like PAD4 [Populus euphratica] |
| 2 |
Hb_000359_080 |
0.1023657915 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 3 |
Hb_000003_420 |
0.1041222522 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_000752_020 |
0.1047987817 |
- |
- |
PREDICTED: uncharacterized protein LOC105641803 isoform X1 [Jatropha curcas] |
| 5 |
Hb_073119_010 |
0.1197111942 |
- |
- |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 6 |
Hb_000103_400 |
0.119713985 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002272_140 |
0.1256245824 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL6-like [Jatropha curcas] |
| 8 |
Hb_000776_050 |
0.12568944 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 9 |
Hb_029866_050 |
0.1264676409 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF4 [Jatropha curcas] |
| 10 |
Hb_002284_020 |
0.1272529998 |
- |
- |
PREDICTED: probable inorganic phosphate transporter 1-9 [Jatropha curcas] |
| 11 |
Hb_000363_110 |
0.1287543277 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 21 [Jatropha curcas] |
| 12 |
Hb_000771_070 |
0.1298100252 |
- |
- |
PREDICTED: plastidial pyruvate kinase 4, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001898_010 |
0.1337814189 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 14 |
Hb_009083_010 |
0.1409072127 |
- |
- |
Phytosulfokines 3 precursor family protein [Populus trichocarpa] |
| 15 |
Hb_000397_090 |
0.1416981258 |
- |
- |
PREDICTED: uncharacterized protein LOC105641235 [Jatropha curcas] |
| 16 |
Hb_000172_080 |
0.1418744084 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 17 |
Hb_009476_110 |
0.1434546673 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 18 |
Hb_000027_130 |
0.1476486567 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 19 |
Hb_000107_440 |
0.1492782076 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A synthase |
hydroxymethylglutaryl-CoA reductase [Hevea brasiliensis] |
| 20 |
Hb_003914_050 |
0.1516667436 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |