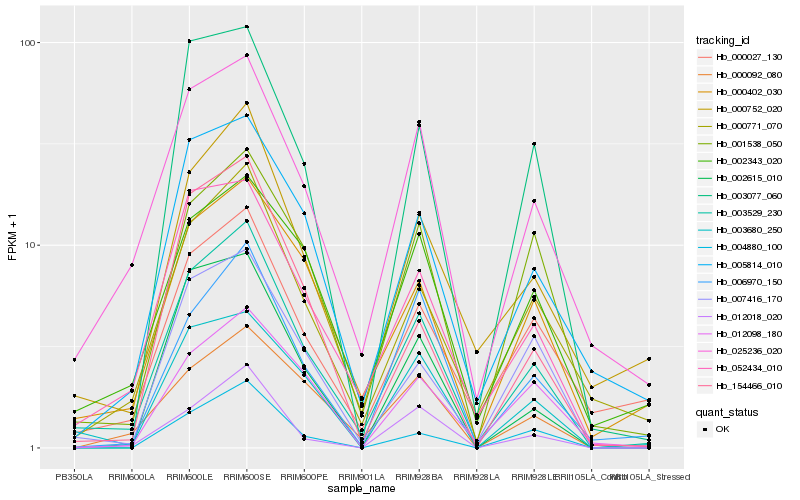
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003529_230 |
0.0 |
transcription factor |
TF Family: ERF |
- |
| 2 |
Hb_052434_010 |
0.1132186632 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like serine/threonine-protein kinase At3g14840 [Sesamum indicum] |
| 3 |
Hb_025236_020 |
0.122330724 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 4 |
Hb_000771_070 |
0.1248565021 |
- |
- |
PREDICTED: plastidial pyruvate kinase 4, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000402_030 |
0.1280943553 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 2 [Jatropha curcas] |
| 6 |
Hb_000752_020 |
0.1307575312 |
- |
- |
PREDICTED: uncharacterized protein LOC105641803 isoform X1 [Jatropha curcas] |
| 7 |
Hb_005814_010 |
0.1330952423 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_003680_250 |
0.1368999368 |
- |
- |
PREDICTED: probable 3-deoxy-D-manno-octulosonic acid transferase, mitochondrial isoform X2 [Jatropha curcas] |
| 9 |
Hb_007416_170 |
0.1382882679 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 10 |
Hb_000027_130 |
0.1465810803 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 11 |
Hb_001538_050 |
0.1476606001 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 12 |
Hb_002343_020 |
0.1489988138 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 13 |
Hb_003077_060 |
0.1512742245 |
- |
- |
PREDICTED: 3-oxo-Delta(4,5)-steroid 5-beta-reductase-like [Jatropha curcas] |
| 14 |
Hb_000092_080 |
0.1524936053 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein VITISV_038742 [Vitis vinifera] |
| 15 |
Hb_154466_010 |
0.1548201857 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 16 |
Hb_012018_020 |
0.157083743 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Eucalyptus grandis] |
| 17 |
Hb_002615_010 |
0.1573534173 |
- |
- |
Uncharacterized protein TCM_040942 [Theobroma cacao] |
| 18 |
Hb_006970_150 |
0.1595431474 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |
| 19 |
Hb_012098_180 |
0.1598778063 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 20 |
Hb_004880_100 |
0.160012018 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11300-like [Glycine max] |