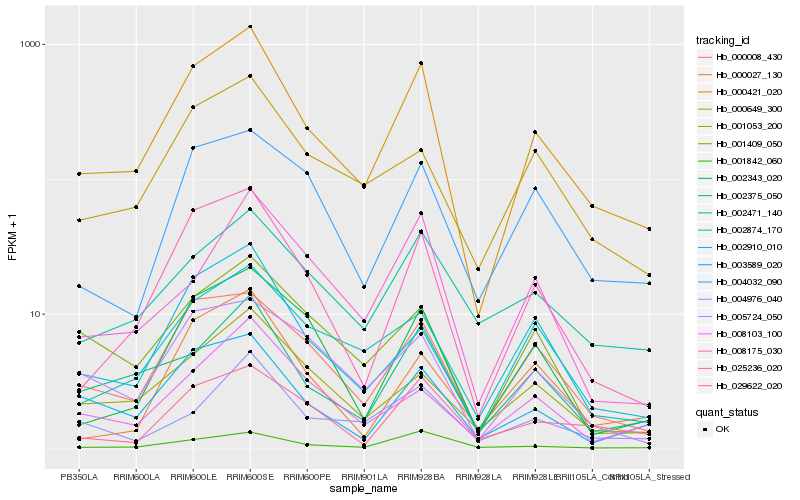
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000421_020 |
0.0 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |
| 2 |
Hb_025236_020 |
0.120357944 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 3 |
Hb_000008_430 |
0.132677434 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 24 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002375_050 |
0.1372675466 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 5 |
Hb_002910_010 |
0.1477619541 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 6 |
Hb_003589_020 |
0.1482006606 |
- |
- |
- |
| 7 |
Hb_002343_020 |
0.1547434926 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 8 |
Hb_000649_300 |
0.1570179444 |
transcription factor |
TF Family: Tify |
JAZ9 [Hevea brasiliensis] |
| 9 |
Hb_001842_060 |
0.1578635112 |
desease resistance |
Gene Name: ABC_trans_N |
hypothetical protein JCGZ_10908 [Jatropha curcas] |
| 10 |
Hb_001053_200 |
0.1588534504 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 11 |
Hb_002471_140 |
0.1625233102 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 12 |
Hb_002874_170 |
0.1639191273 |
- |
- |
PREDICTED: L-arabinokinase-like isoform X3 [Jatropha curcas] |
| 13 |
Hb_005724_050 |
0.1650084168 |
- |
- |
- |
| 14 |
Hb_029622_020 |
0.1673404484 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004032_090 |
0.1686987548 |
- |
- |
PREDICTED: protein AIG1 [Jatropha curcas] |
| 16 |
Hb_001409_050 |
0.1695884393 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_004976_040 |
0.1750981338 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |
| 18 |
Hb_008103_100 |
0.1754142777 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPS2 [Jatropha curcas] |
| 19 |
Hb_000027_130 |
0.1754240169 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 20 |
Hb_008175_030 |
0.1769372979 |
transcription factor |
TF Family: TAZ |
PREDICTED: BTB/POZ and TAZ domain-containing protein 3 [Jatropha curcas] |