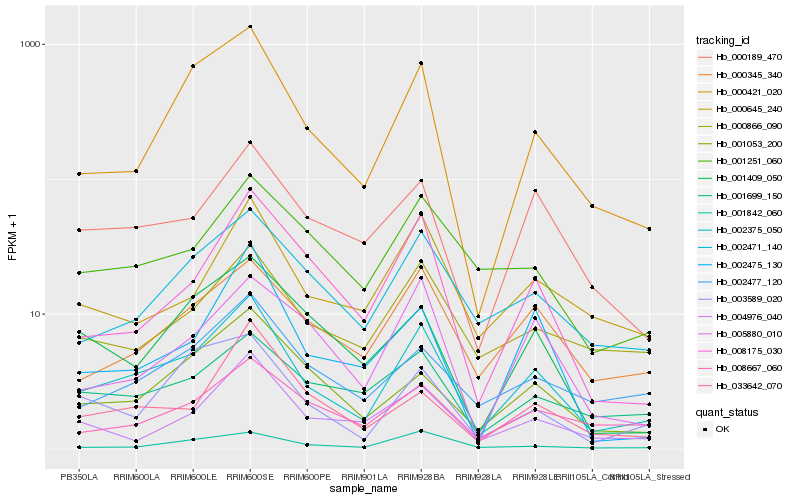
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002375_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 2 |
Hb_000421_020 |
0.1372675466 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |
| 3 |
Hb_000189_470 |
0.1579292374 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT102 [Jatropha curcas] |
| 4 |
Hb_002471_140 |
0.159126026 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 5 |
Hb_000345_340 |
0.1598371022 |
- |
- |
PREDICTED: DNA polymerase zeta catalytic subunit [Jatropha curcas] |
| 6 |
Hb_001053_200 |
0.1674200872 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 7 |
Hb_008175_030 |
0.1681225329 |
transcription factor |
TF Family: TAZ |
PREDICTED: BTB/POZ and TAZ domain-containing protein 3 [Jatropha curcas] |
| 8 |
Hb_002477_120 |
0.1686185727 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 9 |
Hb_000645_240 |
0.1704415041 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 10 |
Hb_000866_090 |
0.1716535006 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X3 [Jatropha curcas] |
| 11 |
Hb_008667_060 |
0.1720872051 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 12 |
Hb_004976_040 |
0.1739073202 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |
| 13 |
Hb_001699_150 |
0.1741626762 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |
| 14 |
Hb_003589_020 |
0.174381058 |
- |
- |
- |
| 15 |
Hb_001842_060 |
0.1757863785 |
desease resistance |
Gene Name: ABC_trans_N |
hypothetical protein JCGZ_10908 [Jatropha curcas] |
| 16 |
Hb_001251_060 |
0.1772012015 |
- |
- |
aldo/keto reductase AKR [Manihot esculenta] |
| 17 |
Hb_005880_010 |
0.1797442363 |
- |
- |
PREDICTED: uncharacterized protein LOC105646431 [Jatropha curcas] |
| 18 |
Hb_033642_070 |
0.1822190655 |
- |
- |
PREDICTED: oligopeptide transporter 7-like [Jatropha curcas] |
| 19 |
Hb_002475_130 |
0.1823050728 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 4 [Jatropha curcas] |
| 20 |
Hb_001409_050 |
0.1833321473 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |