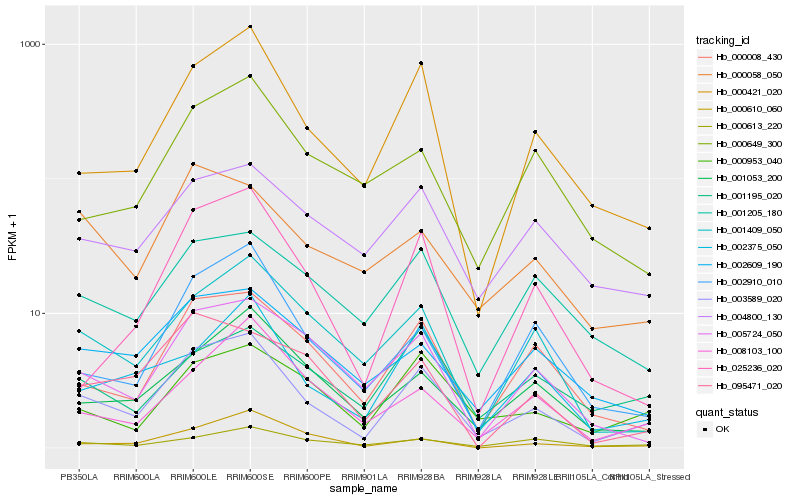
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003589_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000421_020 |
0.1482006606 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |
| 3 |
Hb_000008_430 |
0.1599847839 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 24 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002375_050 |
0.174381058 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 5 |
Hb_000953_040 |
0.174968746 |
- |
- |
hypothetical protein OsJ_19135 [Oryza sativa Japonica Group] |
| 6 |
Hb_002910_010 |
0.1773548058 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 7 |
Hb_001409_050 |
0.1784791345 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_001053_200 |
0.1786515563 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 9 |
Hb_005724_050 |
0.180897411 |
- |
- |
- |
| 10 |
Hb_002609_190 |
0.1817666382 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 11 |
Hb_004800_130 |
0.1834220489 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 12 |
Hb_001205_180 |
0.1895281012 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 13 |
Hb_025236_020 |
0.1895887631 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 14 |
Hb_008103_100 |
0.1898588868 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPS2 [Jatropha curcas] |
| 15 |
Hb_000613_220 |
0.1938195659 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 16 |
Hb_000058_050 |
0.1945711012 |
- |
- |
PREDICTED: uncharacterized protein LOC105640877 [Jatropha curcas] |
| 17 |
Hb_095471_020 |
0.1980495449 |
- |
- |
- |
| 18 |
Hb_000649_300 |
0.2011415977 |
transcription factor |
TF Family: Tify |
JAZ9 [Hevea brasiliensis] |
| 19 |
Hb_001195_020 |
0.2012904222 |
- |
- |
PREDICTED: ornithine carbamoyltransferase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000610_060 |
0.201297795 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |