| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
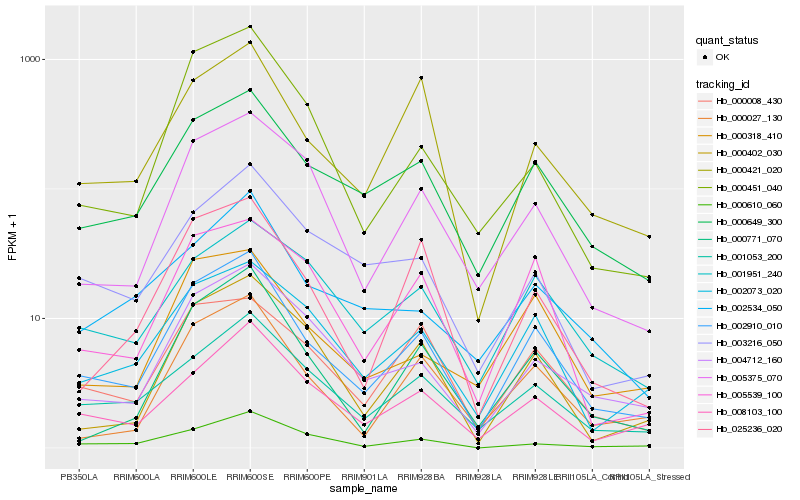
Hb_002910_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 2 |
Hb_000649_300 |
0.10926582 |
transcription factor |
TF Family: Tify |
JAZ9 [Hevea brasiliensis] |
| 3 |
Hb_001053_200 |
0.1280662427 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 4 |
Hb_008103_100 |
0.1307278333 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPS2 [Jatropha curcas] |
| 5 |
Hb_000027_130 |
0.1339838661 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 6 |
Hb_000771_070 |
0.1355724208 |
- |
- |
PREDICTED: plastidial pyruvate kinase 4, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002073_020 |
0.1375084698 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 8 |
Hb_004712_160 |
0.1403266589 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 9 |
Hb_005375_070 |
0.1421386913 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein LHY [Jatropha curcas] |
| 10 |
Hb_000318_410 |
0.1437287263 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1-like [Jatropha curcas] |
| 11 |
Hb_000421_020 |
0.1477619541 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |
| 12 |
Hb_000402_030 |
0.1485353143 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 2 [Jatropha curcas] |
| 13 |
Hb_000008_430 |
0.1486127146 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 24 isoform X1 [Jatropha curcas] |
| 14 |
Hb_025236_020 |
0.1509794902 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 15 |
Hb_005539_100 |
0.1519304758 |
- |
- |
nucleoside transporter, putative [Ricinus communis] |
| 16 |
Hb_000610_060 |
0.1546456259 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 17 |
Hb_003216_050 |
0.1552996446 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 18 |
Hb_000451_040 |
0.1556014553 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002534_050 |
0.1569216161 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |
| 20 |
Hb_001951_240 |
0.1588192521 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |