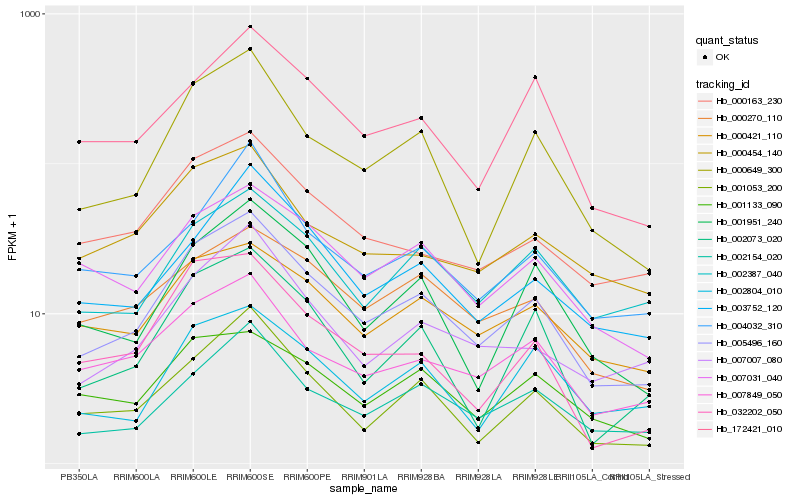
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005496_160 |
0.0 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000649_300 |
0.0879816987 |
transcription factor |
TF Family: Tify |
JAZ9 [Hevea brasiliensis] |
| 3 |
Hb_002154_020 |
0.1033924012 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_007849_050 |
0.108579735 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 5 |
Hb_001053_200 |
0.1094767805 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 6 |
Hb_007007_080 |
0.110954935 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001951_240 |
0.1144642625 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 8 |
Hb_003752_120 |
0.1163856455 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 9 |
Hb_007031_040 |
0.1166571464 |
transcription factor |
TF Family: bZIP |
PREDICTED: bZIP transcription factor 17-like [Jatropha curcas] |
| 10 |
Hb_000163_230 |
0.1168671999 |
- |
- |
Phytochrome A-associated F-box protein, putative [Ricinus communis] |
| 11 |
Hb_000421_110 |
0.1172749129 |
- |
- |
PREDICTED: V-type proton ATPase subunit a1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002073_020 |
0.1190198523 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 13 |
Hb_002387_040 |
0.1196195058 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 14 |
Hb_000270_110 |
0.1197841991 |
- |
- |
PREDICTED: calcium-dependent protein kinase 10 [Jatropha curcas] |
| 15 |
Hb_172421_010 |
0.122055806 |
- |
- |
cysteine protease [Hevea brasiliensis] |
| 16 |
Hb_000454_140 |
0.1239600499 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2a-like [Jatropha curcas] |
| 17 |
Hb_001133_090 |
0.1264616536 |
- |
- |
hypothetical protein POPTR_0012s02680g [Populus trichocarpa] |
| 18 |
Hb_004032_310 |
0.1271289907 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 19 |
Hb_032202_050 |
0.127407826 |
transcription factor |
TF Family: TCP |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002804_010 |
0.1276209644 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like [Jatropha curcas] |