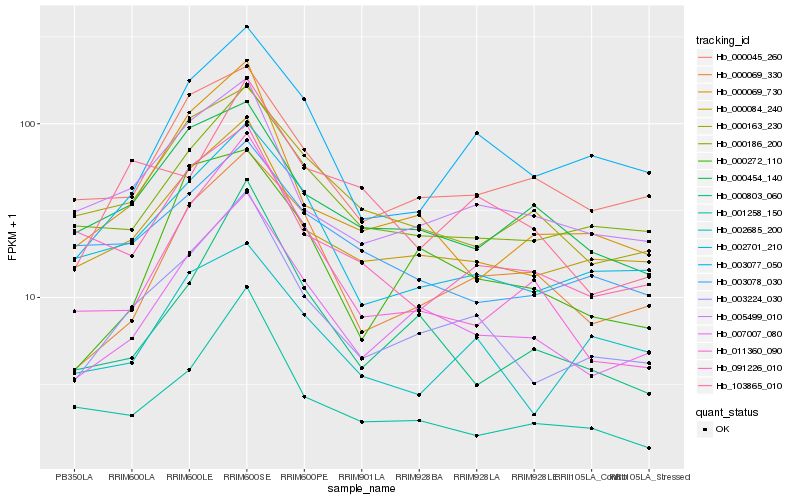
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003224_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0077s002402g, partial [Populus trichocarpa] |
| 2 |
Hb_007007_080 |
0.1043963458 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000084_240 |
0.105260447 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 4 |
Hb_002701_210 |
0.1238191895 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 5 |
Hb_005499_010 |
0.1254915291 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 6 |
Hb_000186_200 |
0.1321204352 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 7 |
Hb_000069_730 |
0.1333314356 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_003077_050 |
0.144198754 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 9 |
Hb_002685_200 |
0.1514630217 |
- |
- |
PREDICTED: uncharacterized protein LOC105632583 [Jatropha curcas] |
| 10 |
Hb_001258_150 |
0.1548355169 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |
| 11 |
Hb_000045_260 |
0.1550166783 |
- |
- |
protein with unknown function [Ricinus communis] |
| 12 |
Hb_000069_330 |
0.1553876986 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_000163_230 |
0.1589504845 |
- |
- |
Phytochrome A-associated F-box protein, putative [Ricinus communis] |
| 14 |
Hb_091226_010 |
0.159315396 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA isoform X1 [Jatropha curcas] |
| 15 |
Hb_103865_010 |
0.1605494157 |
- |
- |
hypothetical protein 21 [Hevea brasiliensis] |
| 16 |
Hb_000454_140 |
0.1621066437 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2a-like [Jatropha curcas] |
| 17 |
Hb_011360_090 |
0.1629923026 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 18 |
Hb_000803_060 |
0.1630648111 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 19 |
Hb_000272_110 |
0.1637000024 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003078_030 |
0.1641316087 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |