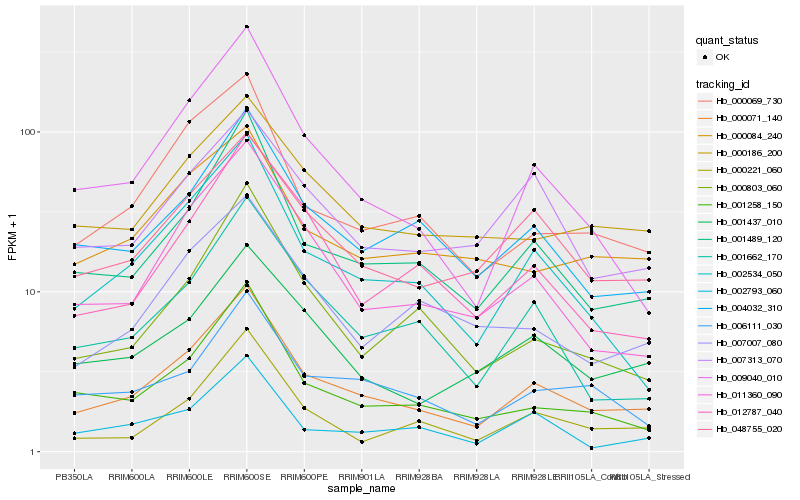
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000071_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628258 [Jatropha curcas] |
| 2 |
Hb_001489_120 |
0.0838134613 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002534_050 |
0.1031792918 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |
| 4 |
Hb_048755_020 |
0.1072756153 |
- |
- |
hypothetical protein JCGZ_09860 [Jatropha curcas] |
| 5 |
Hb_000069_730 |
0.110431037 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_004032_310 |
0.1111115313 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 7 |
Hb_001258_150 |
0.1124942291 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |
| 8 |
Hb_011360_090 |
0.1139123547 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 9 |
Hb_009040_010 |
0.1212932918 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_001437_010 |
0.1228134687 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g33260 [Jatropha curcas] |
| 11 |
Hb_001662_170 |
0.1232070695 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 12 |
Hb_000186_200 |
0.1237753453 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 13 |
Hb_006111_030 |
0.1260141941 |
- |
- |
PREDICTED: phospholipase A(1) LCAT3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000803_060 |
0.1294730619 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 15 |
Hb_012787_040 |
0.1302379073 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000084_240 |
0.1321208314 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 17 |
Hb_002793_060 |
0.1328823763 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g02980 [Populus euphratica] |
| 18 |
Hb_007313_070 |
0.1334089491 |
- |
- |
PREDICTED: gibberellin receptor GID1C [Jatropha curcas] |
| 19 |
Hb_000221_060 |
0.1350185632 |
- |
- |
- |
| 20 |
Hb_007007_080 |
0.1351967096 |
- |
- |
conserved hypothetical protein [Ricinus communis] |