| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
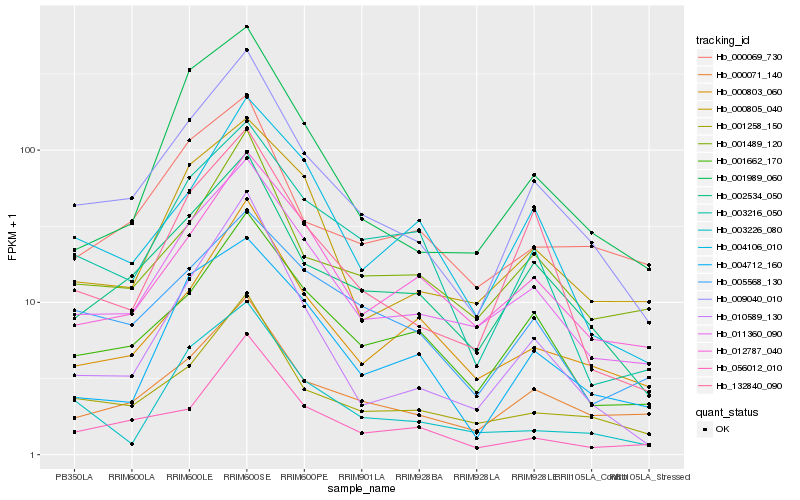
Hb_009040_010 |
0.0 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_002534_050 |
0.089809175 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |
| 3 |
Hb_011360_090 |
0.1030159876 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 4 |
Hb_001258_150 |
0.1184437786 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |
| 5 |
Hb_000071_140 |
0.1212932918 |
- |
- |
PREDICTED: uncharacterized protein LOC105628258 [Jatropha curcas] |
| 6 |
Hb_132840_090 |
0.1230829986 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001662_170 |
0.1268047416 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 8 |
Hb_010589_130 |
0.1328950806 |
- |
- |
PREDICTED: uncharacterized protein LOC104884346 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_001489_120 |
0.1354566629 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001989_060 |
0.1374445261 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105634562 isoform X2 [Jatropha curcas] |
| 11 |
Hb_056012_010 |
0.141623755 |
- |
- |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 12 |
Hb_000803_060 |
0.1449705691 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 13 |
Hb_003216_050 |
0.1471130152 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 14 |
Hb_012787_040 |
0.1479999465 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000805_040 |
0.1486499269 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 16 |
Hb_000069_730 |
0.1516807969 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_004712_160 |
0.1521587479 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 18 |
Hb_004106_010 |
0.1557263386 |
- |
- |
PREDICTED: protein GPR107 isoform X1 [Jatropha curcas] |
| 19 |
Hb_005568_130 |
0.1561457437 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_003226_080 |
0.1588087595 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5-like [Jatropha curcas] |