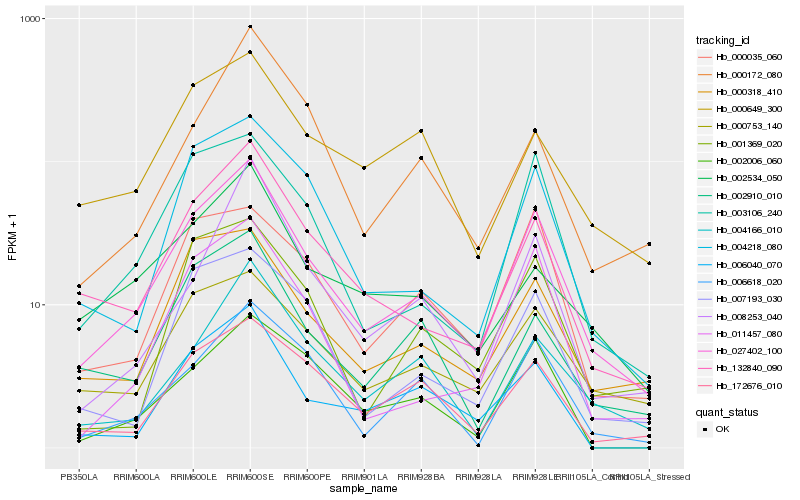
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027402_100 |
0.0 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |
| 2 |
Hb_132840_090 |
0.1237432693 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003106_240 |
0.1323211161 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 4 |
Hb_004218_080 |
0.1373765003 |
- |
- |
PREDICTED: uncharacterized protein LOC105631207 [Jatropha curcas] |
| 5 |
Hb_000753_140 |
0.1383757243 |
- |
- |
PREDICTED: uncharacterized protein LOC105640672 [Jatropha curcas] |
| 6 |
Hb_000318_410 |
0.1414102897 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1-like [Jatropha curcas] |
| 7 |
Hb_011457_080 |
0.1450341907 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_172676_010 |
0.1506233532 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 9 |
Hb_007193_030 |
0.1521493361 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 10 |
Hb_006618_020 |
0.1524214974 |
- |
- |
Vacuolar-processing enzyme precursor [Ricinus communis] |
| 11 |
Hb_004166_010 |
0.1577312363 |
- |
- |
PREDICTED: uncharacterized protein LOC105638508 [Jatropha curcas] |
| 12 |
Hb_001369_020 |
0.1592938357 |
- |
- |
PREDICTED: U-box domain-containing protein 35-like [Jatropha curcas] |
| 13 |
Hb_000035_060 |
0.1597703604 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Jatropha curcas] |
| 14 |
Hb_002910_010 |
0.1599838246 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 15 |
Hb_006040_070 |
0.1621354315 |
- |
- |
orf107b gene product (mitochondrion) [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_008253_040 |
0.1681605998 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 17 |
Hb_002534_050 |
0.1689641742 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |
| 18 |
Hb_002006_060 |
0.1698868282 |
- |
- |
PREDICTED: oleosin 1-like [Jatropha curcas] |
| 19 |
Hb_000649_300 |
0.1738907393 |
transcription factor |
TF Family: Tify |
JAZ9 [Hevea brasiliensis] |
| 20 |
Hb_000172_080 |
0.1744819188 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |