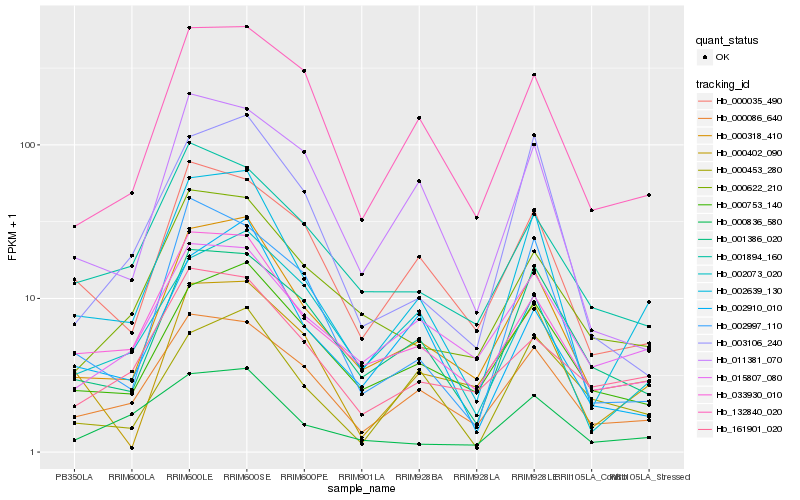
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002639_130 |
0.0 |
- |
- |
hypothetical protein JCGZ_08129 [Jatropha curcas] |
| 2 |
Hb_000318_410 |
0.1159471159 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1-like [Jatropha curcas] |
| 3 |
Hb_033930_010 |
0.1366663392 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 4 |
Hb_000086_640 |
0.1424066038 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 5 |
Hb_000836_580 |
0.1600314665 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000035_490 |
0.1626613831 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 7 |
Hb_002997_110 |
0.1632835168 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 22 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001386_020 |
0.1666233273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_132840_020 |
0.1672701933 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 10 |
Hb_000753_140 |
0.1681234745 |
- |
- |
PREDICTED: uncharacterized protein LOC105640672 [Jatropha curcas] |
| 11 |
Hb_003106_240 |
0.1738241185 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 12 |
Hb_002073_020 |
0.1755589939 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 13 |
Hb_002910_010 |
0.1756352998 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 14 |
Hb_000402_090 |
0.1766466453 |
- |
- |
Lipase [Glycine soja] |
| 15 |
Hb_015807_080 |
0.1782589235 |
- |
- |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |
| 16 |
Hb_000622_210 |
0.1797959451 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X3 [Jatropha curcas] |
| 17 |
Hb_161901_020 |
0.1798814157 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Vitis vinifera] |
| 18 |
Hb_011381_070 |
0.1808156088 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 19 |
Hb_000453_280 |
0.1820728948 |
- |
- |
negative cofactor 2 transcriptional co-repressor, putative [Ricinus communis] |
| 20 |
Hb_001894_160 |
0.1832924744 |
- |
- |
conserved hypothetical protein [Ricinus communis] |