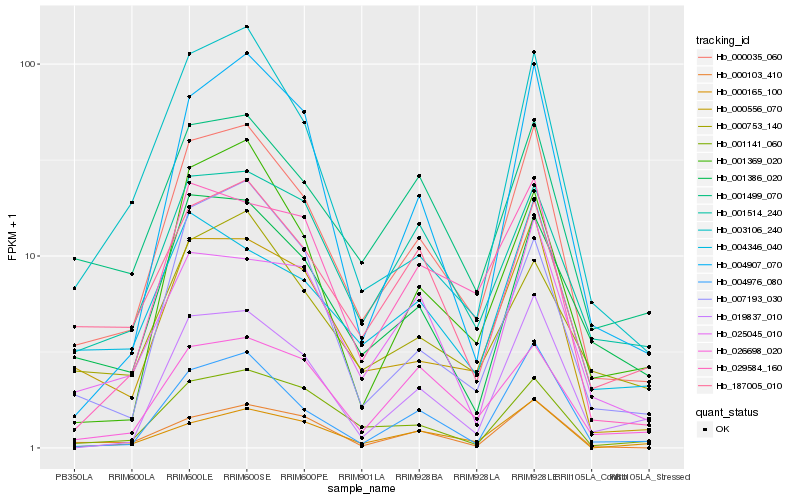
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000035_060 |
0.0 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000556_070 |
0.1061174065 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At2g45590 [Jatropha curcas] |
| 3 |
Hb_001141_060 |
0.1260222159 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001499_070 |
0.1279235716 |
- |
- |
PREDICTED: calcium-transporting ATPase 9, plasma membrane-type isoform X1 [Jatropha curcas] |
| 5 |
Hb_187005_010 |
0.1310127792 |
transcription factor |
TF Family: MYB-related |
PREDICTED: 1-phosphatidylinositol 3-phosphate 5-kinase [Jatropha curcas] |
| 6 |
Hb_004976_080 |
0.131456229 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 7 |
Hb_001386_020 |
0.1330492651 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004907_070 |
0.1340883066 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 9 |
Hb_003106_240 |
0.1348695535 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 10 |
Hb_000103_410 |
0.1350823134 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 11 |
Hb_019837_010 |
0.1380955205 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_029584_160 |
0.1390335064 |
- |
- |
PREDICTED: thioredoxin-like 2, chloroplastic [Populus euphratica] |
| 13 |
Hb_000165_100 |
0.1428945409 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 14 |
Hb_001514_240 |
0.1435716008 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 15 |
Hb_025045_010 |
0.1446341682 |
- |
- |
catalytic, putative [Ricinus communis] |
| 16 |
Hb_026698_020 |
0.1449845776 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 17 |
Hb_001369_020 |
0.1450168175 |
- |
- |
PREDICTED: U-box domain-containing protein 35-like [Jatropha curcas] |
| 18 |
Hb_000753_140 |
0.1462643427 |
- |
- |
PREDICTED: uncharacterized protein LOC105640672 [Jatropha curcas] |
| 19 |
Hb_007193_030 |
0.148415067 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004346_040 |
0.1504786275 |
- |
- |
hypothetical protein POPTR_0016s06630g [Populus trichocarpa] |