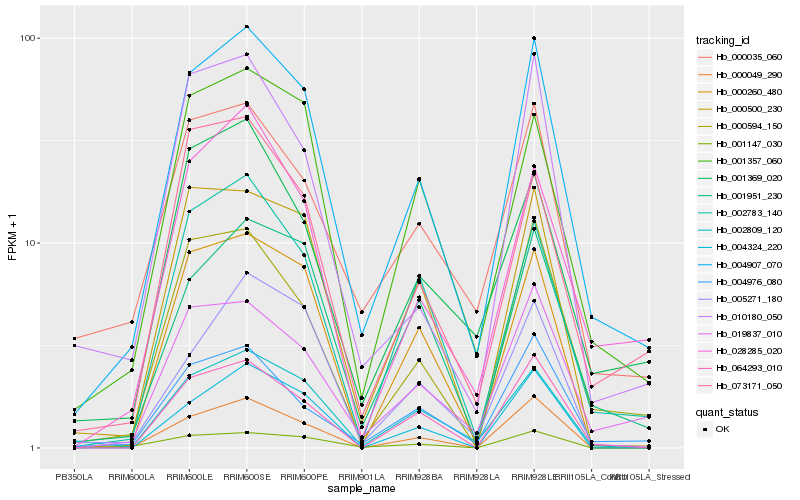
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004907_070 |
0.0 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 2 |
Hb_019837_010 |
0.1133391132 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001951_230 |
0.1158304332 |
- |
- |
PREDICTED: phytochrome E isoform X2 [Jatropha curcas] |
| 4 |
Hb_000594_150 |
0.116928016 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic-like isoform X1 [Nelumbo nucifera] |
| 5 |
Hb_001357_060 |
0.1190547597 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 18-like [Cicer arietinum] |
| 6 |
Hb_002809_120 |
0.1193510891 |
- |
- |
Uncharacterized protein TCM_031281 [Theobroma cacao] |
| 7 |
Hb_004976_080 |
0.120299082 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 8 |
Hb_002783_140 |
0.1228781639 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 9 |
Hb_000049_290 |
0.1293835372 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 10 |
Hb_028285_020 |
0.1312271941 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_000035_060 |
0.1340883066 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Jatropha curcas] |
| 12 |
Hb_064293_010 |
0.1347068985 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 13 |
Hb_000260_480 |
0.1356483292 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 14 |
Hb_005271_180 |
0.1372439591 |
- |
- |
Leucine-rich repeat receptor protein kinase EXS precursor, putative [Ricinus communis] |
| 15 |
Hb_001147_030 |
0.1375366116 |
- |
- |
hypothetical protein JCGZ_14809 [Jatropha curcas] |
| 16 |
Hb_000500_230 |
0.138243678 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 17 |
Hb_073171_050 |
0.1400181773 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 18 |
Hb_001369_020 |
0.1415206375 |
- |
- |
PREDICTED: U-box domain-containing protein 35-like [Jatropha curcas] |
| 19 |
Hb_004324_220 |
0.1430449744 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 20 |
Hb_010180_050 |
0.1433184835 |
- |
- |
PREDICTED: CBL-interacting protein kinase 2 [Jatropha curcas] |