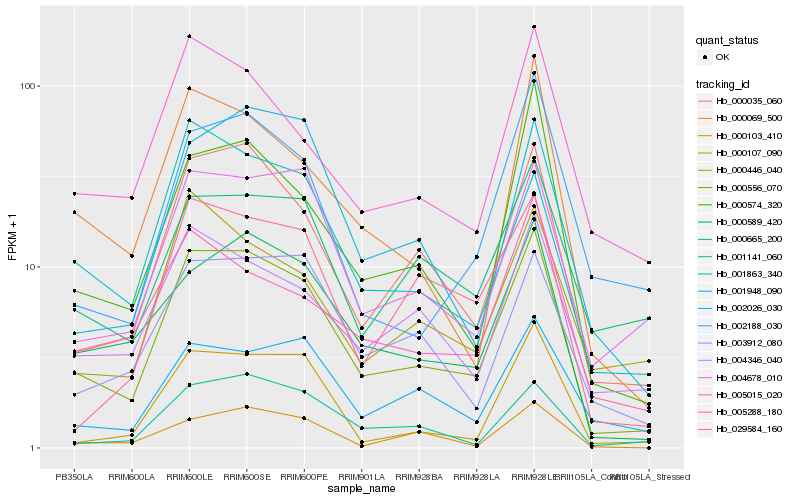
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000556_070 |
0.0 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At2g45590 [Jatropha curcas] |
| 2 |
Hb_000035_060 |
0.1061174065 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000589_420 |
0.1217416807 |
- |
- |
PREDICTED: uncharacterized protein LOC105647273 [Jatropha curcas] |
| 4 |
Hb_004678_010 |
0.1329665686 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 5 |
Hb_000574_320 |
0.1398409783 |
- |
- |
hypothetical protein CICLE_v10024937mg [Citrus clementina] |
| 6 |
Hb_000069_500 |
0.1436781796 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 4, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001141_060 |
0.1444082162 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_004346_040 |
0.1459182771 |
- |
- |
hypothetical protein POPTR_0016s06630g [Populus trichocarpa] |
| 9 |
Hb_003912_080 |
0.1468927386 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 10 |
Hb_005015_020 |
0.1471388874 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_000107_090 |
0.1486399744 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 12 |
Hb_002026_030 |
0.1512090686 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 13 |
Hb_001948_090 |
0.1526584898 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0005s14540g [Populus trichocarpa] |
| 14 |
Hb_029584_160 |
0.1533355845 |
- |
- |
PREDICTED: thioredoxin-like 2, chloroplastic [Populus euphratica] |
| 15 |
Hb_000103_410 |
0.1545229656 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 16 |
Hb_000665_200 |
0.1553812898 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 17 |
Hb_002188_030 |
0.1568926328 |
- |
- |
PREDICTED: probable zinc metallopeptidase EGY3, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_001863_340 |
0.1574829019 |
- |
- |
PREDICTED: probable pectin methyltransferase QUA2 [Jatropha curcas] |
| 19 |
Hb_000446_040 |
0.1592749542 |
- |
- |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 20 |
Hb_005288_180 |
0.1596665445 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04760, chloroplastic isoform X1 [Jatropha curcas] |