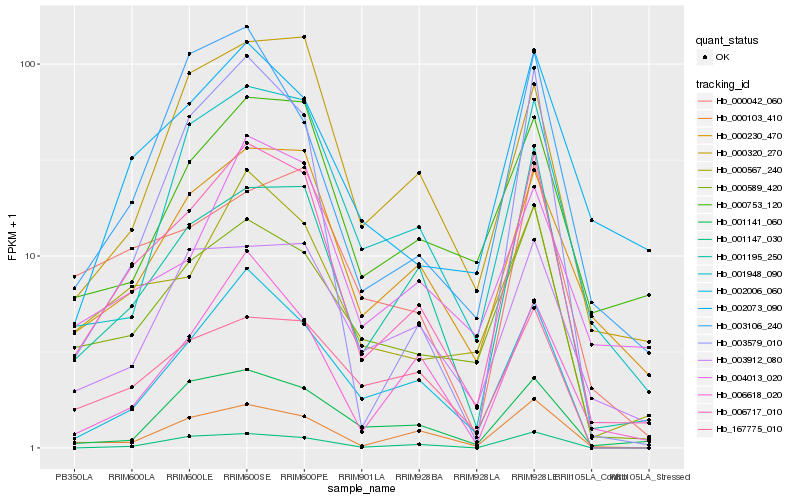
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006717_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649338 [Jatropha curcas] |
| 2 |
Hb_001195_250 |
0.1275038665 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 3 |
Hb_000567_240 |
0.1392374359 |
- |
- |
PREDICTED: F-box/LRR-repeat MAX2 homolog A-like [Jatropha curcas] |
| 4 |
Hb_000103_410 |
0.1425834131 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 5 |
Hb_002006_060 |
0.1425879613 |
- |
- |
PREDICTED: oleosin 1-like [Jatropha curcas] |
| 6 |
Hb_000589_420 |
0.1467462109 |
- |
- |
PREDICTED: uncharacterized protein LOC105647273 [Jatropha curcas] |
| 7 |
Hb_001948_090 |
0.1483933421 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0005s14540g [Populus trichocarpa] |
| 8 |
Hb_000230_470 |
0.1509168473 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 9 |
Hb_003579_010 |
0.1605452142 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 10 |
Hb_000320_270 |
0.1651293733 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002073_090 |
0.1654406592 |
- |
- |
PREDICTED: copper transporter 6-like [Jatropha curcas] |
| 12 |
Hb_006618_020 |
0.1667066023 |
- |
- |
Vacuolar-processing enzyme precursor [Ricinus communis] |
| 13 |
Hb_167775_010 |
0.1692176832 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 14 |
Hb_001147_030 |
0.1694683278 |
- |
- |
hypothetical protein JCGZ_14809 [Jatropha curcas] |
| 15 |
Hb_000753_120 |
0.1716610489 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 16 |
Hb_003912_080 |
0.1726260056 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 17 |
Hb_000042_060 |
0.1737275196 |
- |
- |
PREDICTED: uncharacterized protein LOC105632794 [Jatropha curcas] |
| 18 |
Hb_003106_240 |
0.1751567484 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 19 |
Hb_001141_060 |
0.1754500656 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_004013_020 |
0.1780587301 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |