| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
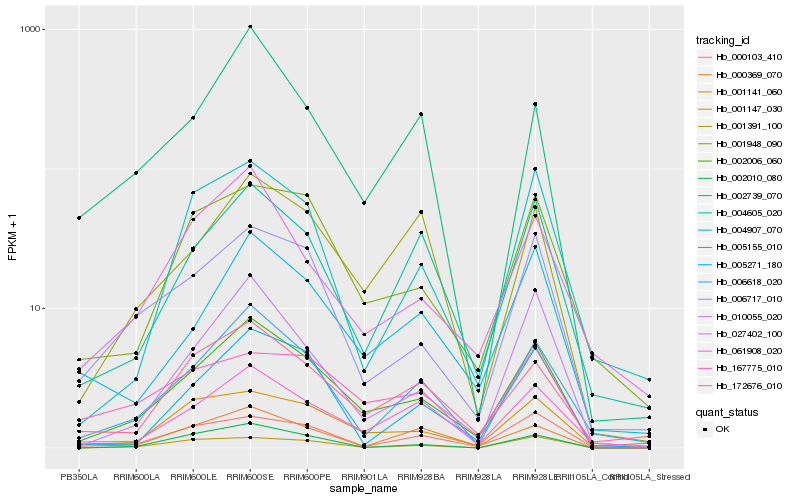
Hb_002006_060 |
0.0 |
- |
- |
PREDICTED: oleosin 1-like [Jatropha curcas] |
| 2 |
Hb_006618_020 |
0.138858581 |
- |
- |
Vacuolar-processing enzyme precursor [Ricinus communis] |
| 3 |
Hb_006717_010 |
0.1425879613 |
- |
- |
PREDICTED: uncharacterized protein LOC105649338 [Jatropha curcas] |
| 4 |
Hb_001948_090 |
0.1481165011 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0005s14540g [Populus trichocarpa] |
| 5 |
Hb_005155_010 |
0.1487875609 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 6 |
Hb_061908_020 |
0.1494493392 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 7 |
Hb_172676_010 |
0.1512189575 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 8 |
Hb_001141_060 |
0.1555927417 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_004605_020 |
0.156014832 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 10 |
Hb_010055_020 |
0.1561546071 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 11 |
Hb_002010_080 |
0.1583728026 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 [Jatropha curcas] |
| 12 |
Hb_001391_100 |
0.1616834147 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 13 |
Hb_005271_180 |
0.1618398909 |
- |
- |
Leucine-rich repeat receptor protein kinase EXS precursor, putative [Ricinus communis] |
| 14 |
Hb_000369_070 |
0.162849014 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 15 |
Hb_000103_410 |
0.1659952189 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 16 |
Hb_001147_030 |
0.168887142 |
- |
- |
hypothetical protein JCGZ_14809 [Jatropha curcas] |
| 17 |
Hb_027402_100 |
0.1698868282 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |
| 18 |
Hb_004907_070 |
0.1733377561 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 19 |
Hb_167775_010 |
0.174657866 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 20 |
Hb_002739_070 |
0.1756602407 |
- |
- |
hypothetical protein B456_002G169900 [Gossypium raimondii] |