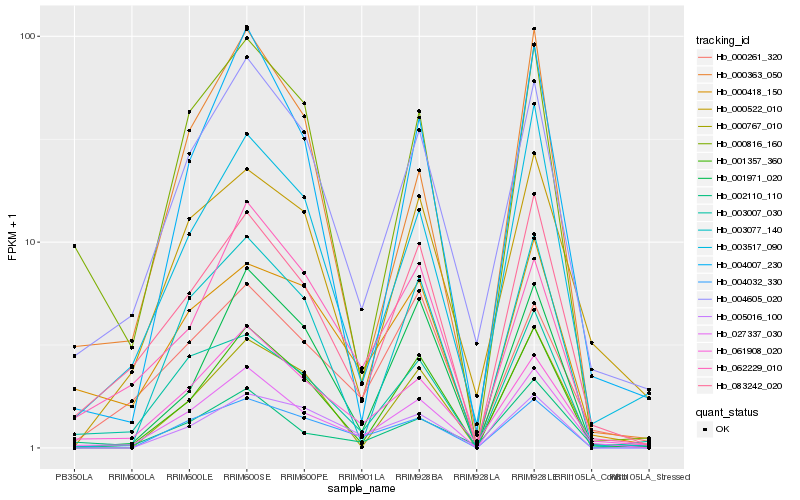
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027337_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104602841 [Nelumbo nucifera] |
| 2 |
Hb_083242_020 |
0.1005135634 |
- |
- |
hypothetical protein JCGZ_20097 [Jatropha curcas] |
| 3 |
Hb_004605_020 |
0.1150734299 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 4 |
Hb_061908_020 |
0.1220288645 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 5 |
Hb_004007_230 |
0.1402000957 |
- |
- |
PREDICTED: putative formamidase C869.04 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004032_330 |
0.1411305117 |
- |
- |
PREDICTED: calvin cycle protein CP12-3, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002110_110 |
0.1412057219 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 8 |
Hb_003517_090 |
0.1415150033 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_003077_140 |
0.1432438222 |
- |
- |
hypothetical protein JCGZ_16294 [Jatropha curcas] |
| 10 |
Hb_003007_030 |
0.1441490368 |
- |
- |
- |
| 11 |
Hb_001357_360 |
0.1443458728 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_000418_150 |
0.1452531662 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_000816_160 |
0.1469631642 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 14 |
Hb_000767_010 |
0.155660834 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like isoform X2 [Solanum lycopersicum] |
| 15 |
Hb_000522_010 |
0.1579252459 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like [Jatropha curcas] |
| 16 |
Hb_005016_100 |
0.1593754728 |
- |
- |
- |
| 17 |
Hb_000261_320 |
0.1611440957 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_000363_050 |
0.1625581597 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_001971_020 |
0.1629406704 |
- |
- |
PREDICTED: alpha-dioxygenase 2 [Populus euphratica] |
| 20 |
Hb_062229_010 |
0.1644416451 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |