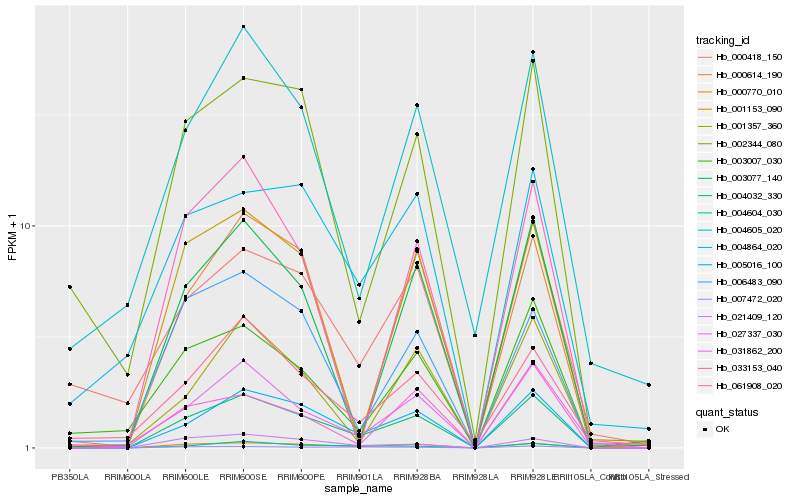
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004032_330 |
0.0 |
- |
- |
PREDICTED: calvin cycle protein CP12-3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_005016_100 |
0.0699432364 |
- |
- |
- |
| 3 |
Hb_000770_010 |
0.0768258575 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 4 |
Hb_021409_120 |
0.1377552944 |
transcription factor |
TF Family: MYB-related |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 5 |
Hb_004604_030 |
0.1405349703 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_027337_030 |
0.1411305117 |
- |
- |
PREDICTED: uncharacterized protein LOC104602841 [Nelumbo nucifera] |
| 7 |
Hb_003077_140 |
0.1512682493 |
- |
- |
hypothetical protein JCGZ_16294 [Jatropha curcas] |
| 8 |
Hb_001153_090 |
0.1568484116 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 9 |
Hb_007472_020 |
0.1616077836 |
- |
- |
hypothetical protein PRUPE_ppa016115mg [Prunus persica] |
| 10 |
Hb_006483_090 |
0.1631236963 |
- |
- |
PREDICTED: uncharacterized protein LOC105639979 [Jatropha curcas] |
| 11 |
Hb_002344_080 |
0.1635446805 |
- |
- |
PREDICTED: protein YLS2-like [Populus euphratica] |
| 12 |
Hb_000614_190 |
0.1653498886 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 13 |
Hb_001357_360 |
0.1674372954 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_061908_020 |
0.1701924423 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 15 |
Hb_004605_020 |
0.1714564173 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 16 |
Hb_031862_200 |
0.1720573094 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 17 |
Hb_004864_020 |
0.1723611334 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 18 |
Hb_000418_150 |
0.1742117089 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_003007_030 |
0.174308954 |
- |
- |
- |
| 20 |
Hb_033153_040 |
0.1751401061 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |