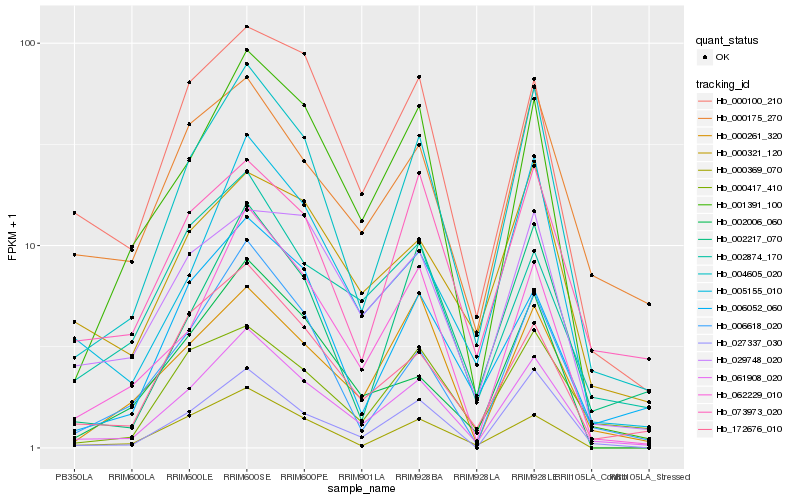
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_061908_020 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 2 |
Hb_004605_020 |
0.0694175787 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 3 |
Hb_062229_010 |
0.1036752718 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 4 |
Hb_001391_100 |
0.116910662 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 5 |
Hb_172676_010 |
0.1178657361 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 6 |
Hb_027337_030 |
0.1220288645 |
- |
- |
PREDICTED: uncharacterized protein LOC104602841 [Nelumbo nucifera] |
| 7 |
Hb_006618_020 |
0.1260294471 |
- |
- |
Vacuolar-processing enzyme precursor [Ricinus communis] |
| 8 |
Hb_005155_010 |
0.1297949073 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 9 |
Hb_000100_210 |
0.1310526457 |
- |
- |
PREDICTED: selenium-binding protein 1-like [Jatropha curcas] |
| 10 |
Hb_000369_070 |
0.1373893271 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 11 |
Hb_000175_270 |
0.1392463348 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000261_320 |
0.1405655869 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_029748_020 |
0.1439948841 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_006052_060 |
0.144157152 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 15 |
Hb_002874_170 |
0.1463669562 |
- |
- |
PREDICTED: L-arabinokinase-like isoform X3 [Jatropha curcas] |
| 16 |
Hb_000417_410 |
0.1473054705 |
- |
- |
PREDICTED: protein STICHEL-like 3 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002217_070 |
0.1482096025 |
- |
- |
PREDICTED: uncharacterized protein LOC105643596 [Jatropha curcas] |
| 18 |
Hb_073973_020 |
0.1482806144 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 19 |
Hb_000321_120 |
0.1491974075 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 20 |
Hb_002006_060 |
0.1494493392 |
- |
- |
PREDICTED: oleosin 1-like [Jatropha curcas] |