| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
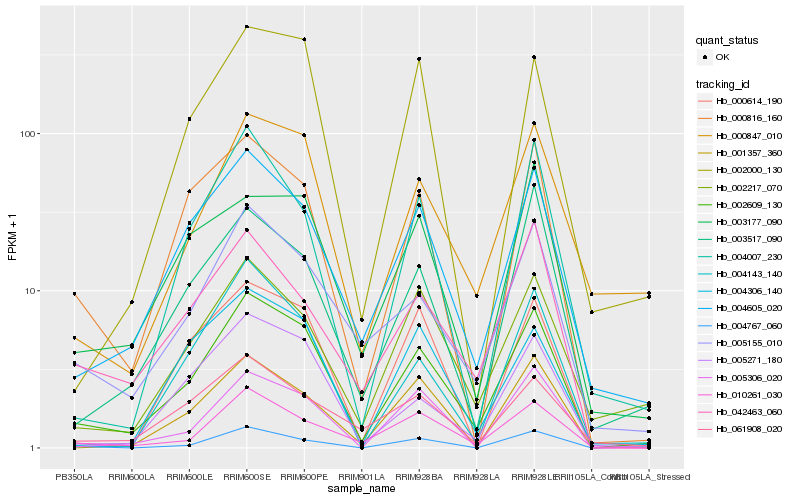
Hb_002609_130 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 86A1 [Jatropha curcas] |
| 2 |
Hb_004605_020 |
0.1221081196 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 3 |
Hb_005155_010 |
0.1339094826 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 4 |
Hb_000847_010 |
0.1390680465 |
- |
- |
PREDICTED: uncharacterized protein LOC102608064 [Citrus sinensis] |
| 5 |
Hb_000816_160 |
0.1395919977 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 6 |
Hb_010261_030 |
0.1402757589 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 7 |
Hb_005271_180 |
0.1416553902 |
- |
- |
Leucine-rich repeat receptor protein kinase EXS precursor, putative [Ricinus communis] |
| 8 |
Hb_002217_070 |
0.1532274763 |
- |
- |
PREDICTED: uncharacterized protein LOC105643596 [Jatropha curcas] |
| 9 |
Hb_004767_060 |
0.1570080522 |
- |
- |
PREDICTED: uncharacterized protein LOC105761928 [Gossypium raimondii] |
| 10 |
Hb_005306_020 |
0.1572826752 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 11 |
Hb_000614_190 |
0.1577608476 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 12 |
Hb_002000_130 |
0.1577699544 |
- |
- |
PREDICTED: abscisic stress-ripening protein 2-like [Eucalyptus grandis] |
| 13 |
Hb_003517_090 |
0.1589388486 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_042463_060 |
0.1603661541 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001357_360 |
0.1604543838 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_004143_140 |
0.160931159 |
- |
- |
glycosyltransferase, partial [Arabidopsis thaliana] |
| 17 |
Hb_004007_230 |
0.1620186712 |
- |
- |
PREDICTED: putative formamidase C869.04 isoform X1 [Jatropha curcas] |
| 18 |
Hb_061908_020 |
0.1624258642 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 19 |
Hb_003177_090 |
0.1633945674 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 20 |
Hb_004306_140 |
0.1638609103 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |