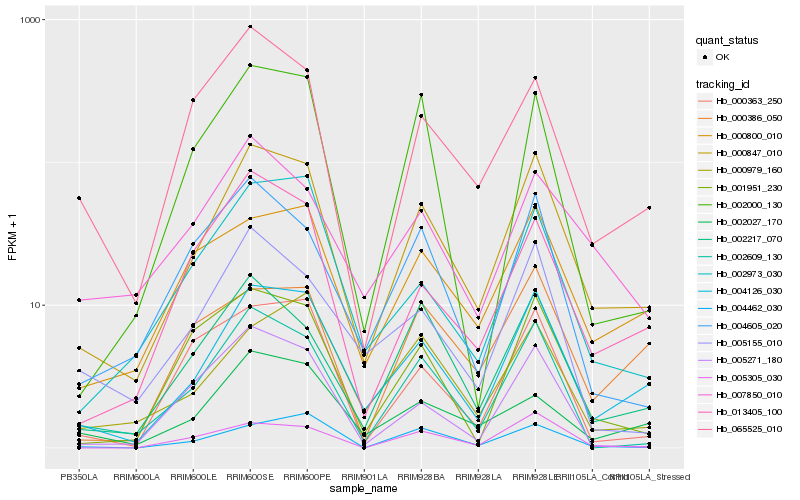
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000847_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC102608064 [Citrus sinensis] |
| 2 |
Hb_004126_030 |
0.0827039548 |
- |
- |
kinase, putative [Ricinus communis] |
| 3 |
Hb_002609_130 |
0.1390680465 |
- |
- |
PREDICTED: cytochrome P450 86A1 [Jatropha curcas] |
| 4 |
Hb_002217_070 |
0.1469541643 |
- |
- |
PREDICTED: uncharacterized protein LOC105643596 [Jatropha curcas] |
| 5 |
Hb_000800_010 |
0.1536940131 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 6 |
Hb_005305_030 |
0.1618589094 |
- |
- |
hypothetical protein JCGZ_25922 [Jatropha curcas] |
| 7 |
Hb_013405_100 |
0.1625510263 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 8 [Jatropha curcas] |
| 8 |
Hb_001951_230 |
0.1645219865 |
- |
- |
PREDICTED: phytochrome E isoform X2 [Jatropha curcas] |
| 9 |
Hb_002973_030 |
0.1660786112 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 10 |
Hb_004605_020 |
0.1679168747 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 11 |
Hb_065525_010 |
0.168060301 |
- |
- |
hypothetical protein JCGZ_01481 [Jatropha curcas] |
| 12 |
Hb_005155_010 |
0.1704664473 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_002000_130 |
0.1774987014 |
- |
- |
PREDICTED: abscisic stress-ripening protein 2-like [Eucalyptus grandis] |
| 14 |
Hb_007850_010 |
0.1788387072 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.4 [Jatropha curcas] |
| 15 |
Hb_002027_170 |
0.1802956154 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RKF3 [Jatropha curcas] |
| 16 |
Hb_005271_180 |
0.1807472417 |
- |
- |
Leucine-rich repeat receptor protein kinase EXS precursor, putative [Ricinus communis] |
| 17 |
Hb_000979_160 |
0.1812553073 |
- |
- |
hypothetical protein RCOM_0629030 [Ricinus communis] |
| 18 |
Hb_000363_250 |
0.1812575376 |
- |
- |
PREDICTED: pirin-like protein [Malus domestica] |
| 19 |
Hb_004462_030 |
0.1820298162 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 20 |
Hb_000386_050 |
0.1831543926 |
- |
- |
PREDICTED: 2-isopropylmalate synthase 2, chloroplastic-like [Jatropha curcas] |