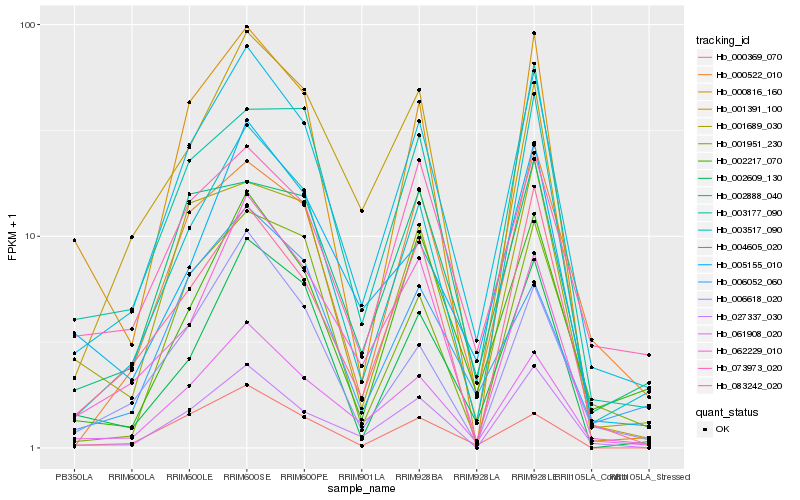
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004605_020 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 2 |
Hb_061908_020 |
0.0694175787 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 3 |
Hb_027337_030 |
0.1150734299 |
- |
- |
PREDICTED: uncharacterized protein LOC104602841 [Nelumbo nucifera] |
| 4 |
Hb_002217_070 |
0.1166395375 |
- |
- |
PREDICTED: uncharacterized protein LOC105643596 [Jatropha curcas] |
| 5 |
Hb_006618_020 |
0.1188956967 |
- |
- |
Vacuolar-processing enzyme precursor [Ricinus communis] |
| 6 |
Hb_002609_130 |
0.1221081196 |
- |
- |
PREDICTED: cytochrome P450 86A1 [Jatropha curcas] |
| 7 |
Hb_062229_010 |
0.1247942265 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 8 |
Hb_000369_070 |
0.1272313205 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 9 |
Hb_000522_010 |
0.1289615682 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like [Jatropha curcas] |
| 10 |
Hb_083242_020 |
0.1305432437 |
- |
- |
hypothetical protein JCGZ_20097 [Jatropha curcas] |
| 11 |
Hb_001391_100 |
0.132403457 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 12 |
Hb_005155_010 |
0.1339559053 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_000816_160 |
0.1341555455 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 14 |
Hb_003517_090 |
0.1353204696 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_001951_230 |
0.1370176728 |
- |
- |
PREDICTED: phytochrome E isoform X2 [Jatropha curcas] |
| 16 |
Hb_001689_030 |
0.1394227075 |
- |
- |
PREDICTED: J domain-containing protein required for chloroplast accumulation response 1 [Jatropha curcas] |
| 17 |
Hb_073973_020 |
0.1414922125 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 18 |
Hb_002888_040 |
0.143265443 |
- |
- |
hypothetical protein B456_009G267200 [Gossypium raimondii] |
| 19 |
Hb_003177_090 |
0.1434942715 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 20 |
Hb_006052_060 |
0.1453027967 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |