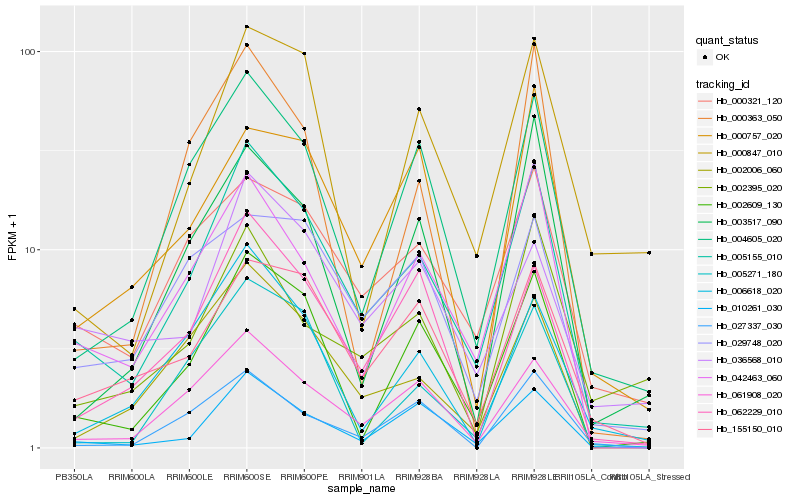
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005155_010 |
0.0 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 2 |
Hb_061908_020 |
0.1297949073 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 3 |
Hb_002609_130 |
0.1339094826 |
- |
- |
PREDICTED: cytochrome P450 86A1 [Jatropha curcas] |
| 4 |
Hb_004605_020 |
0.1339559053 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 5 |
Hb_042463_060 |
0.1353000491 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000321_120 |
0.1405516573 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 7 |
Hb_010261_030 |
0.1422512886 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 8 |
Hb_002006_060 |
0.1487875609 |
- |
- |
PREDICTED: oleosin 1-like [Jatropha curcas] |
| 9 |
Hb_155150_010 |
0.1520696765 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_062229_010 |
0.1624903026 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 11 |
Hb_036568_010 |
0.1663577482 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 23 [Jatropha curcas] |
| 12 |
Hb_006618_020 |
0.1701095497 |
- |
- |
Vacuolar-processing enzyme precursor [Ricinus communis] |
| 13 |
Hb_000847_010 |
0.1704664473 |
- |
- |
PREDICTED: uncharacterized protein LOC102608064 [Citrus sinensis] |
| 14 |
Hb_003517_090 |
0.175777022 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_002395_020 |
0.1781827649 |
- |
- |
Protein CREG1 precursor, putative [Ricinus communis] |
| 16 |
Hb_029748_020 |
0.1784817939 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_027337_030 |
0.1790164101 |
- |
- |
PREDICTED: uncharacterized protein LOC104602841 [Nelumbo nucifera] |
| 18 |
Hb_005271_180 |
0.1797735317 |
- |
- |
Leucine-rich repeat receptor protein kinase EXS precursor, putative [Ricinus communis] |
| 19 |
Hb_000363_050 |
0.180112008 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_000757_020 |
0.1802612823 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |