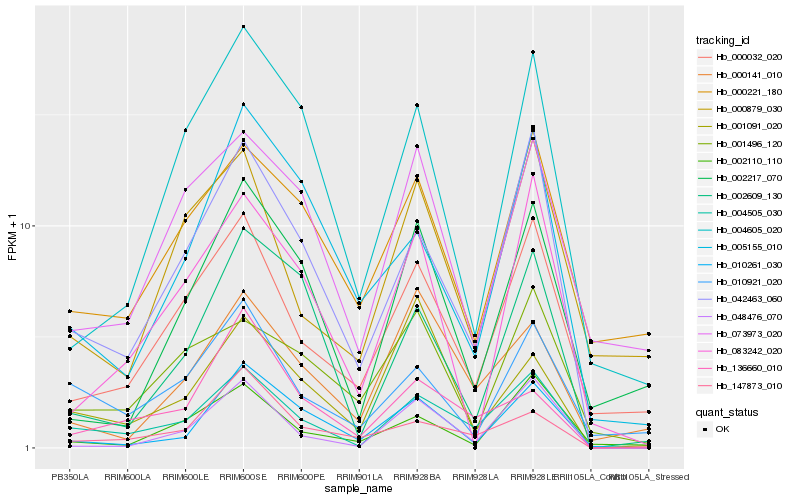
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004505_030 |
0.0 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_042463_060 |
0.1354787075 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000032_020 |
0.1776496178 |
- |
- |
Centromeric protein E, putative [Ricinus communis] |
| 4 |
Hb_010921_020 |
0.1914346174 |
- |
- |
PREDICTED: cyclin-SDS [Jatropha curcas] |
| 5 |
Hb_048476_070 |
0.1978572481 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_010261_030 |
0.2048676809 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 7 |
Hb_002609_130 |
0.2082520962 |
- |
- |
PREDICTED: cytochrome P450 86A1 [Jatropha curcas] |
| 8 |
Hb_147873_010 |
0.2096150904 |
- |
- |
PREDICTED: uncharacterized protein LOC102595311 [Solanum tuberosum] |
| 9 |
Hb_000879_030 |
0.2124315825 |
- |
- |
PREDICTED: uncharacterized protein LOC103707552 [Phoenix dactylifera] |
| 10 |
Hb_000221_180 |
0.2129952581 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 11 |
Hb_004605_020 |
0.2139582316 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 12 |
Hb_000141_010 |
0.2146188427 |
- |
- |
PREDICTED: uncharacterized protein LOC105632253 [Jatropha curcas] |
| 13 |
Hb_073973_020 |
0.2149644305 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 14 |
Hb_136660_010 |
0.2151471587 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_002110_110 |
0.218146506 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 16 |
Hb_005155_010 |
0.2191297001 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 17 |
Hb_083242_020 |
0.2199655153 |
- |
- |
hypothetical protein JCGZ_20097 [Jatropha curcas] |
| 18 |
Hb_001496_120 |
0.2200333503 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_002217_070 |
0.2218153397 |
- |
- |
PREDICTED: uncharacterized protein LOC105643596 [Jatropha curcas] |
| 20 |
Hb_001091_020 |
0.2238398577 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |