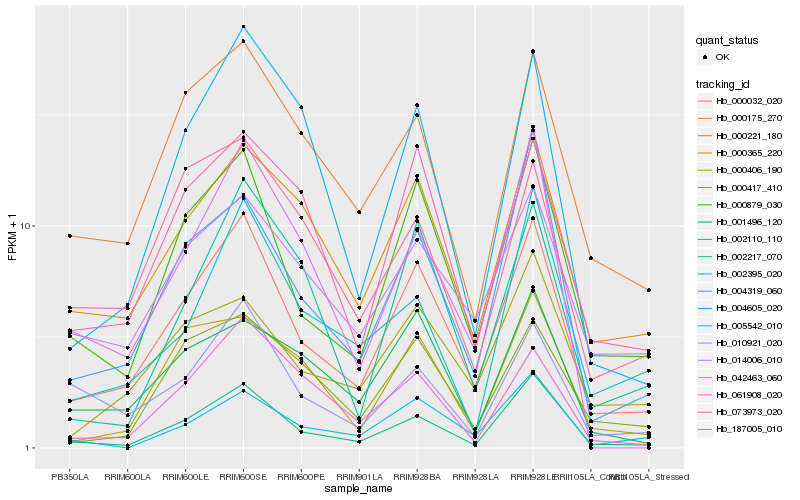
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000032_020 |
0.0 |
- |
- |
Centromeric protein E, putative [Ricinus communis] |
| 2 |
Hb_000879_030 |
0.0838166936 |
- |
- |
PREDICTED: uncharacterized protein LOC103707552 [Phoenix dactylifera] |
| 3 |
Hb_002110_110 |
0.111309878 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 4 |
Hb_000221_180 |
0.1238346607 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 5 |
Hb_073973_020 |
0.1317025226 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 6 |
Hb_000175_270 |
0.1350046982 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 7 |
Hb_005542_010 |
0.1407464573 |
- |
- |
Uncharacterized protein TCM_021447 [Theobroma cacao] |
| 8 |
Hb_014006_010 |
0.1446324209 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 9 |
Hb_000406_190 |
0.144927515 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 10 |
Hb_004605_020 |
0.1514064491 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 11 |
Hb_000417_410 |
0.1520705543 |
- |
- |
PREDICTED: protein STICHEL-like 3 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002217_070 |
0.1540805214 |
- |
- |
PREDICTED: uncharacterized protein LOC105643596 [Jatropha curcas] |
| 13 |
Hb_001496_120 |
0.1558147893 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_061908_020 |
0.1571364693 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 15 |
Hb_010921_020 |
0.1573854652 |
- |
- |
PREDICTED: cyclin-SDS [Jatropha curcas] |
| 16 |
Hb_000365_220 |
0.1573946546 |
- |
- |
protein phosphatase-1, putative [Ricinus communis] |
| 17 |
Hb_042463_060 |
0.1588372007 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004319_060 |
0.158894789 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 19 |
Hb_187005_010 |
0.1659864495 |
transcription factor |
TF Family: MYB-related |
PREDICTED: 1-phosphatidylinositol 3-phosphate 5-kinase [Jatropha curcas] |
| 20 |
Hb_002395_020 |
0.166146049 |
- |
- |
Protein CREG1 precursor, putative [Ricinus communis] |