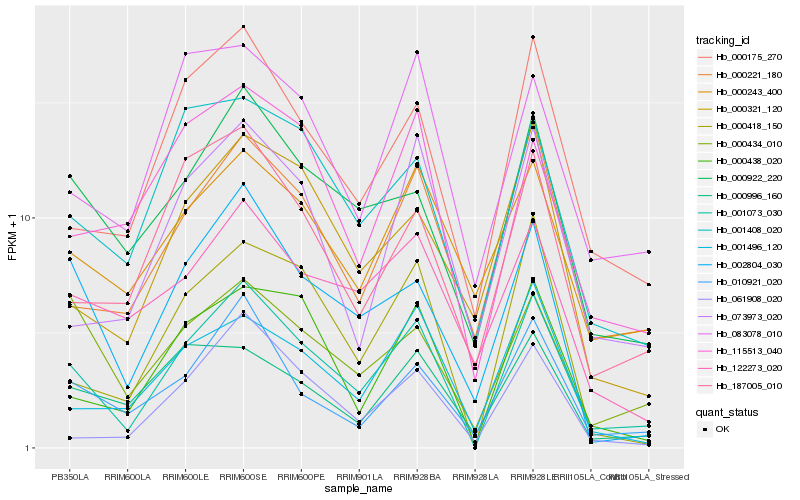
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001073_030 |
0.0 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000175_270 |
0.1454339939 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000438_020 |
0.1497672109 |
- |
- |
leucine-rich repeat-containing protein 2, lrrc2, putative [Ricinus communis] |
| 4 |
Hb_000418_150 |
0.1517043892 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_010921_020 |
0.1527634224 |
- |
- |
PREDICTED: cyclin-SDS [Jatropha curcas] |
| 6 |
Hb_000221_180 |
0.1564529765 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 7 |
Hb_000922_220 |
0.1574592095 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM-like [Jatropha curcas] |
| 8 |
Hb_002804_030 |
0.158469006 |
- |
- |
PREDICTED: probable protein phosphatase 2C 4 [Jatropha curcas] |
| 9 |
Hb_001408_020 |
0.1662892282 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 10 |
Hb_000434_010 |
0.1678763412 |
- |
- |
PREDICTED: protein starmaker [Jatropha curcas] |
| 11 |
Hb_187005_010 |
0.1678885063 |
transcription factor |
TF Family: MYB-related |
PREDICTED: 1-phosphatidylinositol 3-phosphate 5-kinase [Jatropha curcas] |
| 12 |
Hb_001496_120 |
0.1718213677 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_061908_020 |
0.1741021423 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 14 |
Hb_073973_020 |
0.1744269697 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 15 |
Hb_000321_120 |
0.1762315695 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 16 |
Hb_083078_010 |
0.1764496564 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 17 |
Hb_115513_040 |
0.1766665792 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000243_400 |
0.1769037653 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 19 |
Hb_122273_020 |
0.1787621705 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP2 [Jatropha curcas] |
| 20 |
Hb_000996_160 |
0.1790081801 |
- |
- |
conserved hypothetical protein [Ricinus communis] |