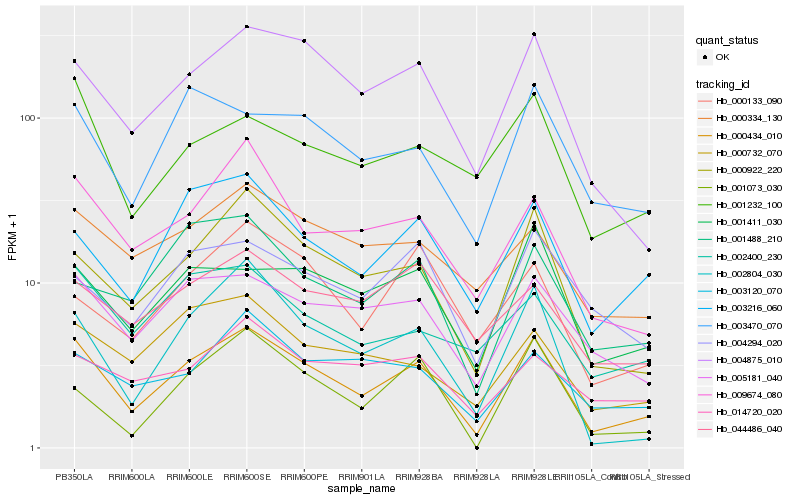
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000434_010 |
0.0 |
- |
- |
PREDICTED: protein starmaker [Jatropha curcas] |
| 2 |
Hb_000922_220 |
0.1181789552 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM-like [Jatropha curcas] |
| 3 |
Hb_004875_010 |
0.1275923994 |
- |
- |
plasma membrane aquaporin 1 [Hevea brasiliensis] |
| 4 |
Hb_005181_040 |
0.1401118411 |
- |
- |
PREDICTED: MAP kinase kinase MKK1/SSP32-like [Jatropha curcas] |
| 5 |
Hb_009674_080 |
0.1407993422 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 6 |
Hb_002804_030 |
0.1431498458 |
- |
- |
PREDICTED: probable protein phosphatase 2C 4 [Jatropha curcas] |
| 7 |
Hb_003216_060 |
0.1433236469 |
- |
- |
PREDICTED: heat shock protein 83 [Jatropha curcas] |
| 8 |
Hb_001411_030 |
0.1452848741 |
- |
- |
hypothetical protein POPTR_0003s09620g [Populus trichocarpa] |
| 9 |
Hb_003470_070 |
0.1526294095 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 10 |
Hb_000732_070 |
0.1578383263 |
- |
- |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 11 |
Hb_000334_130 |
0.1607203955 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 12 |
Hb_003120_070 |
0.1621915563 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_001232_100 |
0.1635191722 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 14 |
Hb_001488_210 |
0.163644186 |
- |
- |
PREDICTED: protein RIK [Jatropha curcas] |
| 15 |
Hb_002400_230 |
0.1658065443 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 6-like [Jatropha curcas] |
| 16 |
Hb_004294_020 |
0.1660174101 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000133_090 |
0.1668075704 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |
| 18 |
Hb_014720_020 |
0.1675021494 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM [Jatropha curcas] |
| 19 |
Hb_001073_030 |
0.1678763412 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 20 |
Hb_044486_040 |
0.1678946343 |
- |
- |
PREDICTED: ion channel CASTOR-like [Malus domestica] |