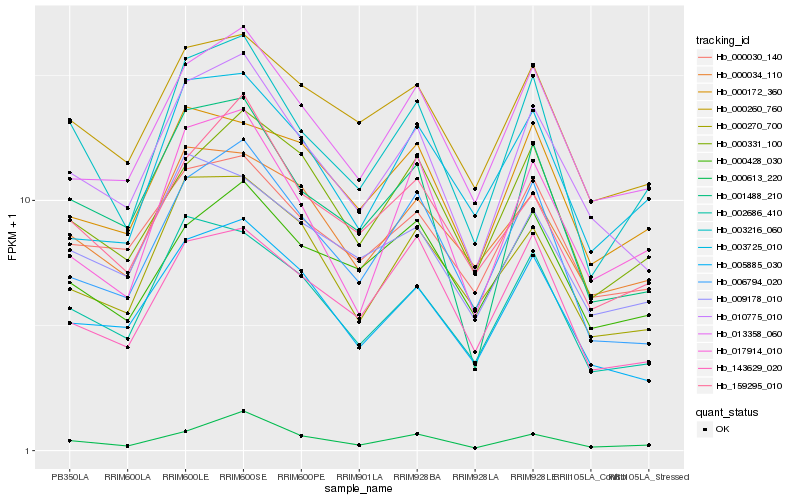
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003216_060 |
0.0 |
- |
- |
PREDICTED: heat shock protein 83 [Jatropha curcas] |
| 2 |
Hb_002686_410 |
0.0931112051 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 48 kDa protein-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_001488_210 |
0.0974215434 |
- |
- |
PREDICTED: protein RIK [Jatropha curcas] |
| 4 |
Hb_010775_010 |
0.0993978683 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000260_760 |
0.1003914747 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 6 |
Hb_006794_020 |
0.1017846428 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 7 |
Hb_013358_060 |
0.1040218879 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 8 |
Hb_159295_010 |
0.1046956835 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 9 |
Hb_000331_100 |
0.1053335426 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 10 |
Hb_017914_010 |
0.1074600327 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 11 |
Hb_000270_700 |
0.1094819194 |
- |
- |
PREDICTED: protease Do-like 9 [Populus euphratica] |
| 12 |
Hb_000030_140 |
0.1095173915 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000428_030 |
0.1098841723 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 14 |
Hb_143629_020 |
0.1099715874 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 15 |
Hb_005885_030 |
0.111115575 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 16 |
Hb_009178_010 |
0.112561277 |
- |
- |
PREDICTED: uncharacterized protein LOC105637474 [Jatropha curcas] |
| 17 |
Hb_000034_110 |
0.1132462318 |
- |
- |
sentrin/sumo-specific protease, putative [Ricinus communis] |
| 18 |
Hb_000613_220 |
0.1145227981 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 19 |
Hb_000172_360 |
0.116849793 |
- |
- |
PREDICTED: apoptotic chromatin condensation inducer in the nucleus [Jatropha curcas] |
| 20 |
Hb_003725_010 |
0.1189025396 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |